Curcumin synthase
| Curcumin synthase 1 (CURS1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.3.1.217 | ||||||||
| CAS number | 1245303-08-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
| Curcumin synthase 2 (CURS2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.3.1.219 | ||||||||
| CAS number | 1245303-09-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
| Curcumin synthase 3 (CURS3) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.3.1.219 | ||||||||
| CAS number | 1245303-10-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
Curcumin synthase categorizes three enzyme isoforms (CURS1, 2, and 3), type III polyketide synthases (PKSs) present in the leaves and rhizome of the turmeric plant (Curcuma longa) [1] that synthesize curcumin.[2] CURS1-3 are responsible for the hydrolysis of feruloyldiketide-CoA,[3] previously produced in the curcuminoid pathway, and a decarboxylative condensation reaction [1][2] that together comprise one of the final steps in the synthesis pathway for curcumin, demethoxycurcumin, and bisdemethoxycurcumin, the compounds that give turmeric both its distinctive yellow color, and traditional medical benefits.[4] CURS should not be confused with Curcuminoid Synthase (CUS), which catalyzes the one-pot synthesis of bisdemethoxycurcumin in Oryza sativa.[5]
Structure
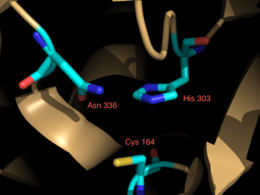
Crystallization studies [6] have determined that curcumin synthase is a homodimer of ketosynthase subunits.[2][7] Each includes a highly conserved Cys (164), His (303), Asn (336) catalytic triad, and CURS1 has been shown to exhibit the αβαβα folding pattern,[6] conserved features of type III PKSs.[7][8] The catalytic triads are independent of each other and are contained in the center of each monomer, connected to the surface with a CoA binding tunnel.[6] While CURS1, 2, and 3 share approximately 80% amino acid sequence identity, their small structural differences account for their differences in preferred starter substrates and most prolific product.[1]
Mechanism
Each CURS catalyzes the reactions necessary to convert a feruloyldiketide-CoA into a curcuminoid, but the three isoforms have preferred starter substrates and products. CURS1 converts feruloyldiketide-CoA esters into curcumin using feruloyl-CoA exclusively as a starter substrate. CURS2 produces both curcumin and demethoxycurcumin, favoring feruloyl-CoA as a starter, and CURS3 produces curcumin, demethoxycurcumin, and bisdemethoxycurcumin from either feruloyl-CoA or 4-coumaroyl-CoA as the starter substrate.[3] The fact that preferences of starter substrates vary between the three CURS is corroborated by carbon labeling studies confirming the incorporation of a variety of starter substrates into curcuminoid products in C. longa.[9]
Only the mechanism of CURS1 has been elucidated. In the first step, the feruloyl moiety of feruloyl-CoA is transferred to Cys (164) followed by feruloyldiketide-CoA entering the CoA binding tunnel and being hydrolyzed through an unknown mechanism to a β-keto acid.[6] The acid is then used as an extender substrate in the catalytic triad, where it undergoes decarboxylative condensation with the feruloyl moiety on Cys (164). This mechanism is thought to be identical to that of the decarboxylative condensation of malonyl-CoA in other type III PKSs.[6] The hydrolysis of the diketide has been shown to be the rate-limiting step of the enzyme.[6]
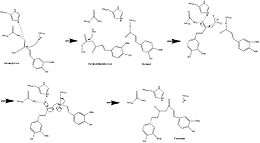
It was previously hypothesized that the curcumoid pathway employed two cinnamoyl-CoAs and one malonyl-CoA, but this was suggested against by the absence of a necessary intermediate such a pathway (bisdeshydroxybisdesmethoxycurcumin),[9] strengthening evidence for feruloyl-CoA or 4-coumaroyl-CoA as starter substrate in CURS.
Biological Activity
The production of curcumin and its derivatives by CURS may be a defense mechanism of C. longa against internal and external threats. Curcumin is a potent antioxidant, as its phenolic structure, highest in activity in curcumin rather than its demethoxylated derivatives,[10] acts as a free-radical scavenging apparatus, eliminating free superoxides and DPPH from the plant’s cells.[10] Curcumin synthase may also protect Curcuma longa from herbivores to some degree, as curcumin has a distinctively bitter taste:[10] studies show CURS1, 2 have higher expression in the leaves of C. longa than the rhizome [1][11] while CURS3 shows equal expression in both locations.[1]
Role in Cancer Research
Research suggests that curcumin is an active anti-cancer molecule against cancers of brain, breast, bones, blood, gastrointestinal tract, genitourinary tract, as well as thoracic and gynecological cancers.[12] The molecule achieves this wide-range activity by up or down-regulating numerous receptors, kinases, growth factors, transcriptional factors, and inflammatory cytokines, among others,[12] thus its biosynthesis is of great interest to medicine.
For instance, curcumin inhibits mammalian nuclear factor κB (NF-κB) by preventing its translocation to the nucleus.[10] This inhibitory action upregulates the levels of preapoptotic and apoptotic cells, eliminating damaged cells, and discouraging abnormal growth patterns, as well as decreasing chemokine levels.[13] As activated NF-κB is associated with oxidative stress,[13] inhibition of the nuclear factor by curcumin is consistent with the chemical’s role as an antioxidant. A homologous system to NF-κB signaling exists in plants,[14] evidence that curcumin may play a similar role in C. longa as it does in humans.
Curcumin synthases in C. longa have been until recently, the only readily available synthesis method of curcumin. Today, laboratory syntheses are capable of producing the chemical,[15] and numerous teams are constructing curcumin analogues designed to target specific biological processes, such as the NFκB signaling pathway previously discussed.[16]
References
- 1 2 3 4 5 Katsuyama, Y.; Kita, T.; Horinouchi, S. (2009). "Identification and characterization of multiple curcumin synthases from the herb Curcuma longa". FEBS Letters. 583 (17): 2799–2803. doi:10.1016/j.febslet.2009.07.029.
- 1 2 3 Katsuyama, Y.; Kita, T.; Funa, N.; Horinouchi, S. (2009). "Curcuminoid Biosynthesis by Two Type III Polyketide Synthases in the Herb Curcuma longa". Journal of Biological Chemistry. 284 (17): 11160–11170. doi:10.1074/jbc.M900070200.
- 1 2 Yu, D.; Xu, F.; Zeng, J.; Zhan, J. (2012). "Type III polyketide synthases in natural product biosynthesis". IUBMB Life. 64 (4): 285–295. doi:10.1002/iub.1005.
- ↑ The Agronomy and Economy of Turmeric and Ginger, K.P. Prabhakaran Nair, The Agronomy and Economy of Turmeric and Ginger: The Invaluable Medicinal Spice Crops, published March 2013, Elsevier. Accessed February 26, 2014 via ScienceDirect.
- ↑ Katsuyama, Y.; Matsuzawa, M.; Funa, N.; Horinouchi, S. (2007). "In Vitro Synthesis of Curcuminoids by Type III Polyketide Synthase from Oryza sativa". Journal of Biological Chemistry. 282 (52): 37702–37709. doi:10.1074/jbc.M707569200.
- 1 2 3 4 5 6 Katsuyama, Y.; Miyazono, K. -I.; Tanokura, M.; Ohnishi, Y.; Horinouchi, S. (2010). "Structural and Biochemical Elucidation of Mechanism for Decarboxylative Condensation of -Keto Acid by Curcumin Synthase". Journal of Biological Chemistry. 286 (8): 6659–68. doi:10.1074/jbc.M110.196279. PMC 3057783
 . PMID 21148316.
. PMID 21148316. - 1 2 Jez, J. M.; Ferrer, J. -L.; Bowman, M. E.; Austin, M. B.; Schröder, J.; Dixon, R. A.; Noel, J. P. (2001). "Structure and mechanism of chalcone synthase-like polyketide synthases". Journal of Industrial Microbiology and Biotechnology. 27 (6): 393–398. doi:10.1038/sj.jim.7000188.
- ↑ Austin, M. B.; Noel, J. P. (2003). "The chalcone synthase superfamily of type III polyketide synthases". Natural Product Reports. Royal Society of Chemistry. 20: 79–110. doi:10.1039/B100917F.
- 1 2 Kita, T.; Imai, S.; Sawada, H.; Kumagai, H.; Seto, H. (2008). "The Biosynthetic Pathway of Curcuminoid in Turmeric (Curcuma longa) as Revealed by 13C-Labeled Precursors". Bioscience, Biotechnology, and Biochemistry. 72 (7): 1789–1798. doi:10.1271/bbb.80075.
- 1 2 3 4 Esatbeyoglu, T.; Huebbe, P.; Ernst, I. M. A.; Chin, D.; Wagner, A. E.; Rimbach, G. (2012). "Curcumin-From Molecule to Biological Function". Angewandte Chemie International Edition. 51 (22): 5308–5332. doi:10.1002/anie.201107724.
- ↑ Ramirez-Ahumada, M. D. C.; Timmermann, B. N.; Gang, D. R. (2006). "Biosynthesis of curcuminoids and gingerols in turmeric (Curcuma longa) and ginger (Zingiber officinale): Identification of curcuminoid synthase and hydroxycinnamoyl-CoA thioesterases". Phytochemistry. 67 (18): 2017–2029. doi:10.1016/j.phytochem.2006.06.028.
- 1 2 Anand, P.; Sundaram, C.; Jhurani, S.; Kunnumakkara, A. B.; Aggarwal, B. B. (2008). "Curcumin and cancer: An "old-age" disease with an "age-old" solution". Cancer Letters. 267: 133–164. doi:10.1016/j.canlet.2008.03.025.
- 1 2 Caamano, J.; Hunter, C. A. (2002). "NF- B Family of Transcription Factors: Central Regulators of Innate and Adaptive Immune Functions". Clinical Microbiology Reviews. 15 (3): 414–429. doi:10.1128/CMR.15.3.414-429.2002.
- ↑ Zhang, G.; Ghosh, S. (2001). "Toll-like receptor–mediated NF-κB activation: A phylogenetically conserved paradigm in innate immunity". Journal of Clinical Investigation. 107: 13–19. doi:10.1172/JCI11837.
- ↑ Babu, K. V. D.; Rajasekharan, K. N. (1994). "Simplified Condition for Synthesis of Curcumin I and Other Curcuminoids". Organic Preparations and Procedures International. 26 (6): 674–677. doi:10.1080/00304949409458165.
- ↑ Qiu, X.; Du, Y.; Lou, B.; Zuo, Y.; Shao, W.; Huo, Y.; Huang, J.; Yu, Y.; Zhou, B.; Du, J.; Fu, H.; Bu, X. (2010). "Synthesis and Identification of New 4-Arylidene Curcumin Analogues as Potential Anticancer Agents Targeting Nuclear Factor-κB Signaling Pathway". Journal of Medicinal Chemistry. 53 (23): 8260–8273. doi:10.1021/jm1004545.