Glutamate—prephenate aminotransferase
| Glutamate-prephenate aminotransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.6.1.79 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
In enzymology, glutamate-prephenate aminotransferase (EC 2.6.1.79, also known as prephenate transaminase, PAT, and L-glutamate:prephenate aminotransferase) is an enzyme that catalyzes the chemical reaction
- L-arogenate + 2-oxoglutarate prephenate + L-glutamate
Thus, the two substrates of this enzyme are L-arogenate and 2-oxoglutarate, whereas its two products are prephenate and L-glutamate. However, in most plant species utilizing this enzyme, the left side of the reaction is strongly favored. Therefore, glutamate is used as the amino donor to convert prephenate into arogenate.
Nomenclature
This enzyme belongs to the family of transferases, specifically the transaminases, which transfer nitrogenous groups. The systematic name of this enzyme class is L-arogenate:2-oxoglutarate aminotransferase. Other names in common use include prephenate transaminase (ambiguous), PAT (ambiguous), and L-glutamate:prephenate aminotransferase. It operates in the phenylalanine and tyrosine biosynthesis pathway.
Species distribution
The gene which encodes this enzyme has recently been identified in various plant species and microorganisms, meaning that all genes in the pathway have now been identified and accounted for. This pathway occurs in many different plant species. As phenylalanine is an essential amino acid, humans (and other animals) have lost the ability to produce it themselves and must therefore obtain it from their diet. As such, the activity of this enzyme in various plant species affects the survival of animals as well. In these animals, tyrosine is synthesized from phenylalanine via the enzyme phenylalanine hydroxylase, whereas plants have their own method of tyrosine synthesis.
Function
Glutamate-Prephanate Aminotransferase catalyzes the reversible reaction shown below:
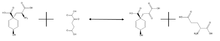 ,
,
and its primary purpose is to convert prephenate into arogenate via transamination, using glutamate as the amino donor. As stated previously, the left side of the reaction is strongly favored. This is a necessary process for any organism which needs to convert arogenate into phenylalanine or tyrosine, as arogenate is an intermediate in the reactions which synthesize these amino acids, an alternative route to that involving phenylpyruvate and hydroxyphenylpyruvate. In the absence of glutamate, aspartate can act as the amino donor in the reaction without the need for a different enzyme, but this reaction proceeds more slowly. The details of the activity of this enzyme are still somewhat of a mystery.
Structure
Little is known about the structure of glutamate-prephenate aminotransferase. However, some data indicates that the enzyme may have an α2-β2 subunit structure.
References and Further reading
- Bonner CA, Jensen RA (1985). "Novel features of prephenate aminotransferase from cell cultures of Nicotiana silvestris". Arch. Biochem. Biophys. 238 (1): 237–46. doi:10.1016/0003-9861(85)90161-4. PMID 3985619.
- Bonner C, Jensen R (1987). "Prephenate aminotransferase". Methods Enzymol. 142: 479–87. doi:10.1016/S0076-6879(87)42059-4. PMID 3298985.
- Siehl DL, Connelly JA, Conn EE (1986). "Tyrosine biosynthesis in Sorghum bicolor: characteristics of prephenate aminotransferase". Z. Naturforsch. C. 41 (1-2): 79–86. PMID 2939644.
- Graindorge M, Giustini C, Jacomin AC, Kraut A, Curien G, Matringe M (2010). "Identification of a plant gene encoding glutamate/aspartate-prephenate aminotransferase: the last homeless enzyme of aromatic amino acids biosynthesis". FEBS Lett. 584 (20): 4357–60. doi:10.1016/j.febslet.2010.09.037. PMID 20883697.
- Maeda H, Yoo H, Dudareva N (2011). "Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate". Nat. Chem. Biol. 7 (1): 19–21. doi:10.1038/nchembio.485. PMID 21102469.
- De-Eknamkul W, Ellis BE (1988). "Purification and characterization of prephenate aminotransferase from Anchusa officinalis cell cultures". Arch. Biochem. Biophys. 267 (1): 87–94. doi:10.1016/0003-9861(88)90011-2. PMID 3196038.
- Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RC, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, Haring MA, Giovannoni J, Fernie AR, Klee HJ (2011). "Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit". Plant Cell. 23 (7): 2738–53. doi:10.1105/tpc.111.086975. PMC 3226207
 . PMID 21750236.
. PMID 21750236. - Graindorge M, Giustini C, Kraut A, Moyet L, Curien G, Matringe M (2014). "Three different classes of aminotransferases evolved prephenate aminotransferase functionality in arogenate-competent microorganisms". J. Biol. Chem. 289 (6): 3198–208. doi:10.1074/jbc.M113.486480. PMID 24302739.
- "Orthology: K15849." KEGG. KEGG. Web. <http://www.kegg.jp/dbget-bin/www_bget?K158492.6.1.79 R07276>.