Structure and genome of HIV
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983.[1][2] Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself did not occur until two years after the first major cases of AIDS associated illnesses were reported in 1981.[3][4]
Structure
HIV is different in structure from other retroviruses. It is around 120 nm in diameter (around 60 times smaller than a red blood cell) and roughly spherical.
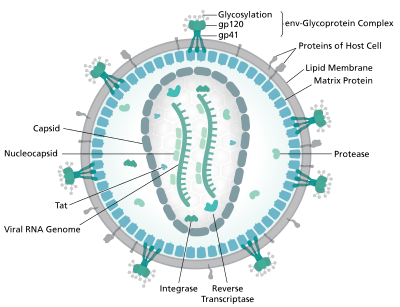
HIV-1 is composed of two copies of noncovalently linked, unspliced, positive-sense single-stranded RNA enclosed by a conical capsid composed of the viral protein p24, typical of lentiviruses.[5][6] The RNA component is 9749 nucleotides long[7][8] and bears a 5’ cap (Gppp), a 3’ poly(A) tail, and many open reading frames (ORFs).[9] Viral structural proteins are encoded by long ORFs, whereas smaller ORFs encode regulators of the viral life cycle: attachment, membrane fusion, replication, and assembly.[9]

The single-strand RNA is tightly bound to p7 nucleocapsid proteins, late assembly protein p6, and enzymes essential to the development of the virion, such as reverse transcriptase and integrase. Lysine tRNA is the primer of the magnesium-dependent reverse transcriptase.[5] The nucleocapsid associates with the genomic RNA (one molecule per hexamer) and protects the RNA from digestion by nucleases. Also enclosed within the virion particle are Vif, Vpr, Nef, and viral protease. A matrix composed of an association of the viral protein p17 surrounds the capsid, ensuring the integrity of the virion particle. This is in turn surrounded by an envelope of host-cell origin. The envelope is formed when the capsid buds from the host cell, taking some of the host-cell membrane with it. The envelope includes the glycoproteins gp120 and gp41, which are responsible for binding to and entering the host cell.
As the only proteins on the surface of the virus, the envelope glycoproteins (gp120 and gp41) are the major targets for HIV vaccine efforts.[10] Over half of the mass of the trimeric envelope spike is N-linked glycans. The density is high as the glycans shield the underlying viral protein from neutralisation by antibodies. This is one of the most densely glycosylated molecules known and the density is sufficiently high to prevent the normal maturation process of glycans during biogenesis in the endoplasmic and Golgi apparatus.[11][12] The majority of the glycans are therefore stalled as immature 'high-mannose' glycans not normally present on secreted or cell surface human glycoproteins.[13] The unusual processing and high density means that almost all broadly neutralising antibodies that have so far been identified (from a subset of patients that have been infected for many months to years) bind to or, are adapted to cope with, these envelope glycans.[14]
The molecular structure of the viral spike has now been determined by X-ray crystallography[15] and cryo-electron microscopy.[16] These advances in structural biology were made possible due to the development of stable recombinant forms of the viral spike by the introduction of an intersubunit disulphide bond and an isoleucine to proline mutation in gp41.[17] The so-called SOSIP trimers not only reproduce the antigenic properties of the native viral spike but also display the same degree of immature glycans as presented on the native virus.[18] Recombinant trimeric viral spikes are promising vaccine candidates as they display less non-neutralising epitopes than recombinant monomeric gp120 which act to suppress the immune response to target epitopes.[19]
Genome organization
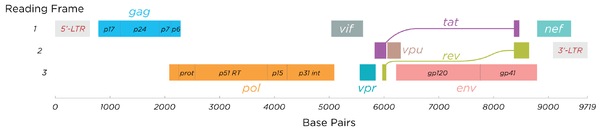
HIV has several major genes coding for structural proteins that are found in all retroviruses as well as several nonstructural ("accessory") genes unique to HIV. The HIV genome contains three major genes, 5'gag-pol-env-3', encoding major structural proteins as well as essential enzymes.[20] These are synthesized as polyproteins which produce proteins for virion interior, called Gag, group specific antigen; the viral enzymes (Pol, polymerase) or the glycoproteins of the virion env (envelope).[21] In addition to these, HIV encodes for proteins which have certain regulatory and auxiliary functions as well.[21] HIV-1 has two important regulatory elements: Tat and Rev and few important accessory proteins such as Nef, Vpr, Vif and Vpu which are not essential for replication in certain tissues.[22] The gag gene provides the basic physical infrastructure of the virus, and pol provides the basic mechanism by which retroviruses reproduce, while the others help HIV to enter the host cell and enhance its reproduction. Though they may be altered by mutation, all of these genes except tev exist in all known variants of HIV; see Genetic variability of HIV.
HIV employs a sophisticated system of differential RNA splicing to obtain nine different gene products from a less than 10kb genome.[23] HIV has a 9.2kb unspliced genomic transcript which encodes for gag and pol precursors; a singly spliced, 4.5 kb encoding for env, Vif, Vpr and Vpu and a multiply spliced, 2 kb mRNA encoding for Tat, Rev and Nef.[23]
| Class | Gene name | Primary protein products | Processed protein products |
|---|---|---|---|
| Viral structural proteins | gag | Gag polyprotein | MA, CA, SP1, NC, SP2, P6 |
| pol | Pol polyprotein | RT, RNase H, IN, PR | |
| env | gp160 | gp120, gp41 | |
| Essential regulatory elements | tat | Tat | |
| rev | Rev | ||
| Accessory regulatory proteins | nef | Nef | |
| vpr | Vpr | ||
| vif | Vif | ||
| vpu | Vpu |
Viral structural proteins
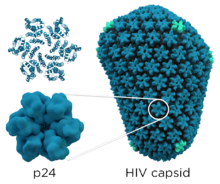
- gag (group-specific antigen) codes for the precursor gag polyprotein which is processed by viral protease during maturation to MA (matrix protein, p17); CA (capsid protein, p24); SP1 (spacer peptide 1, p2); NC (nucleocapsid protein, p7); SP2 (spacer peptide 2, p1) and P6 protein.[24]
- pol codes for viral enzymes reverse transcriptase (RT) and RNase H, integrase (IN), and HIV protease (PR).[21] HIV protease is required to cleave the precursor Gag polyprotein to produce structural proteins, RT is required to transcribe DNA from RNA template, and IN is necessary to integrate the double-stranded viral DNA into the host genome.[20]
- env (for "envelope") codes for gp160, which is cleaved by a host protease, furin, within the endoplasmic reticulum of the host cell. The post-translational processing produces a surface glycoprotein, gp120 or SU, which attaches to the CD4 receptors present on lymphocytes, and gp41 or TM, which embeds in the viral envelope to enable the virus to attach to and fuse with target cells.[20][24]
Essential regulatory elements
- tat (HIV trans-activator) plays an important role in regulating the reverse transcription of viral genome RNA, ensuring efficient synthesis of viral mRNAs and regulating the release of virions from infected cells.[21] Tat is expressed as 72-amino acid one-exon Tat as well as the 86-101 amino-acid two-exon Tat, and plays an important role early in HIV infection. Tat (14-15kDa) binds to the bulged genomic RNA stem-loop secondary structure near the 5' LTR region forming the trans-activation response element (TAR).[5][21]
- rev (regulator of expression of virion proteins): The Rev protein binds to the viral genome via an arginine-rich RNA-binding motif that also acts as a NLS (nuclear localization signals), required for the transport of Rev to the nucleus from cytosol during viral replication.[21] Rev recognizes a complex stem-loop structure of the mRNA env located in the intron separating coding exon of Tat and Rev, known as the HIV Rev response element (RRE).[5][21] Rev is important for the synthesis of major viral proteins and is hence essential for viral replication.
Accessory regulatory proteins
- vpr (lentivirus protein R): Vpr is a virion-associated, nucleocytoplasmic shuttling regulatory protein.[21] It is believed to play an important role in replication of the virus, specifically, nuclear import of the preintegration complex. Vpr also appears to cause its host cells to arrest their cell cycle in the G2 phase. This arrest activates the host DNA repair machinery which may enable integration of the viral DNA.[5] HIV-2 and SIV encode an additional Vpr related protein called Vpx which functions in association with Vpr.[21]
- vif - Vif is a highly conserved, 23 kDa phosphoprotein important for the infectivity of HIV-1 virions depending on the cell type.[5] HIV-1 has been found to require Vif to synthesize infectious viruses in lymphocytes, macrophages, and certain human cell lines. It does not appear to require Vif for the same process in HeLa cells or COS cells, among others.[21]
- nef- Nef, negative factor, is a N-terminal myristoylated membrane-associated phosphoprotein. It is involved in multiple functions during the replication cycle of the virus. It is believed to play an important role in cell apoptosis and increase virus infectivity.[21]
- vpu (Virus protein U) - Vpu is specific to HIV-1. It is a class I oligomeric integral membrane phosphoprotein with numerous biological functions. Vpu is involved in CD4 degradation involving the ubiquitin proteasome pathway as well as in the successful release of virions from infected cells.[5][21]
- tev: This gene is only present in a few HIV-1 isolates. It is a fusion of parts of the tat, env, and rev genes, and codes for a protein with some of the properties of tat, but little or none of the properties of rev.[25]
RNA secondary structure
| HIV pol-1 stem loop | |
|---|---|
 | |
| Predicted secondary structure of the HIV pol-1 stem loop | |
| Identifiers | |
| Symbol | pol |
| Rfam | RF01418 |
| Other data | |
| RNA type | Cis-reg |
Several conserved secondary structure elements have been identified within the HIV RNA genome. The 5'UTR structure consists of series of stem-loop structures connected by small linkers.[6] These stem-loops (5' to 3') include the trans-activation region (TAR) element, the 5' polyadenylation signal [poly(A)], the PBS, the DIS, the major SD and the ψ hairpin structure located within the 5' end of the genome and the HIV Rev response element (RRE) within the env gene.[6][26][27] Another RNA structure that has been identified is gag stem loop 3 (GSL3), thought to be involved in viral packaging.[28][29] RNA secondary structures have been proposed to affect the HIV life cycle by altering the function of HIV protease and reverse transcriptase, although not all elements identified have been assigned a function.
An RNA secondary structure determined by SHAPE analysis has shown to contain three stem loops and is located between the HIV protease and reverse transcriptase genes. This cis regulatory RNA has been shown to be conserved throughout the HIV family and is thought to influence the viral life cycle.[30]
The complete sequence of the HIV-1 genome, extracted from infectious virions, has been solved to single-nucleotide resolution.[31]
See also
References
- ↑ Barré-Sinoussi F, Chermann JC, Rey F, et al. (May 1983). "Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS)". Science. 220 (4599): 868–71. Bibcode:1983Sci...220..868B. doi:10.1126/science.6189183. PMID 6189183.
- ↑ Gallo RC, Sarin PS, Gelmann EP, et al. (May 1983). "Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS)". Science. 220 (4599): 865–7. Bibcode:1983Sci...220..865G. doi:10.1126/science.6601823. PMID 6601823.
- ↑ Centers for Disease Control and Prevention (1981-06-05). "Pneumocycstis Pneumonia – Los Angeles" (PDF). Morbidity and Mortality Weekly Report. 30 (21): 250–2. PMID 6265753. Retrieved 2008-05-10.
- ↑ Centers for Disease Control and Prevention (1981-07-04). "Kaposi's Sarcoma and Pneumocycstis Pneumonia Among Homosexual Men – New York City and California" (PDF). Morbidity and Mortality Weekly Report. 30 (25): 305–8. PMID 6789108. Retrieved 2008-05-10.
- 1 2 3 4 5 6 7 Montagnier, Luc. (1999) Human Immunodeficiency Viruses (Retroviridae). Encyclopedia of Virology (2nd Ed.) 763-774
- 1 2 3 Lu, K; Heng, X; Summers, MF (2011). "Structural determinants and mechanism of HIV-1 genome packaging". Journal of Molecular Biology. 410 (4): 609–33. doi:10.1016/j.jmb.2011.04.029. PMC 3139105
 . PMID 21762803.
. PMID 21762803.
- ↑ Wain-Hobson S, Sonigo P, Danos O, et al. (1985). "Nucleotide sequence of the AIDS virus, LAV". Cell. 40 (1): 9–17. doi:10.1016/0092-8674(85)90303-4. PMID 2981635.
- ↑ Ratner L, Haseltine W, Patarca R, et al. (1985). "Complete nucleotide sequence of the AIDS virus, HTLV-III". Nature. 313 (6000): 277–84. Bibcode:1985Natur.313..277R. doi:10.1038/313277a0. PMID 2578615.
- 1 2 Castelli, Joann C. and Levy, Jay A. (2002) HIV (Human Immunodeficiency Virus). Encyclopedia of Cancer (2nd Ed.) 2:407--415
- ↑ National Institute of Health (June 17, 1998). "Crystal structure of key HIV protein reveals new prevention, treatment targets" (Press release). Archived from the original on February 19, 2006. Retrieved September 14, 2006.
- ↑ Behrens, Anna-Janina; Vasiljevic, Snezana; Pritchard, Laura K.; Harvey, David J.; Andev, Rajinder S.; Krumm, Stefanie A.; Struwe, Weston B.; Cupo, Albert; Kumar, Abhinav (2016-03-10). "Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein". Cell Reports. 0 (0). doi:10.1016/j.celrep.2016.02.058. ISSN 2211-1247.
- ↑ Pritchard, Laura K.; Spencer, Daniel I. R.; Royle, Louise; Bonomelli, Camille; Seabright, Gemma E.; Behrens, Anna-Janina; Kulp, Daniel W.; Menis, Sergey; Krumm, Stefanie A. (2015-06-24). "Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies". Nature Communications. 6: 7479. doi:10.1038/ncomms8479. PMC 4500839
 . PMID 26105115.
. PMID 26105115. - ↑ Pritchard, Laura K.; Harvey, David J.; Bonomelli, Camille; Crispin, Max; Doores, Katie J. (2015-09-01). "Cell- and Protein-Directed Glycosylation of Native Cleaved HIV-1 Envelope". Journal of Virology. 89 (17): 8932–8944. doi:10.1128/JVI.01190-15. ISSN 0022-538X. PMC 4524065
 . PMID 26085151.
. PMID 26085151. - ↑ Crispin, Max; Doores, Katie J (2015-04-01). "Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design". Current Opinion in Virology. Viral pathogenesis • Preventive and therapeutic vaccines. 11: 63–69. doi:10.1016/j.coviro.2015.02.002.
- ↑ Julien, Jean-Philippe; Cupo, Albert; Sok, Devin; Stanfield, Robyn L.; Lyumkis, Dmitry; Deller, Marc C.; Klasse, Per-Johan; Burton, Dennis R.; Sanders, Rogier W. (2013-12-20). "Crystal structure of a soluble cleaved HIV-1 envelope trimer". Science (New York, N.Y.). 342 (6165): 1477–1483. doi:10.1126/science.1245625. ISSN 1095-9203. PMC 3886632
 . PMID 24179159.
. PMID 24179159. - ↑ Lyumkis, Dmitry; Julien, Jean-Philippe; de Val, Natalia; Cupo, Albert; Potter, Clinton S.; Klasse, Per-Johan; Burton, Dennis R.; Sanders, Rogier W.; Moore, John P. (2013-12-20). "Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer". Science (New York, N.Y.). 342 (6165): 1484–1490. doi:10.1126/science.1245627. ISSN 1095-9203. PMC 3954647
 . PMID 24179160.
. PMID 24179160. - ↑ Sanders, Rogier W.; Derking, Ronald; Cupo, Albert; Julien, Jean-Philippe; Yasmeen, Anila; de Val, Natalia; Kim, Helen J.; Blattner, Claudia; de la Peña, Alba Torrents (2013-09-01). "A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies". PLoS Pathogens. 9 (9): e1003618. doi:10.1371/journal.ppat.1003618. ISSN 1553-7374. PMC 3777863
 . PMID 24068931.
. PMID 24068931. - ↑ Pritchard, Laura K.; Vasiljevic, Snezana; Ozorowski, Gabriel; Seabright, Gemma E.; Cupo, Albert; Ringe, Rajesh; Kim, Helen J.; Sanders, Rogier W.; Doores, Katie J. (2015-06-16). "Structural Constraints Determine the Glycosylation of HIV-1 Envelope Trimers". Cell Reports. 11 (10): 1604–1613. doi:10.1016/j.celrep.2015.05.017. ISSN 2211-1247. PMC 4555872
 . PMID 26051934.
. PMID 26051934. - ↑ de Taeye, Steven W.; Ozorowski, Gabriel; Torrents de la Peña, Alba; Guttman, Miklos; Julien, Jean-Philippe; van den Kerkhof, Tom L. G. M.; Burger, Judith A.; Pritchard, Laura K.; Pugach, Pavel (2015-12-17). "Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes". Cell. 163 (7): 1702–1715. doi:10.1016/j.cell.2015.11.056. ISSN 1097-4172. PMC 4732737
 . PMID 26687358.
. PMID 26687358. - 1 2 3 Mushahwar, Isa K. (2007) Human Immunodeficiency Viruses: Molecular Virology, pathogenesis, diagnosis and treatment. Perspectives in Medical Virology. 13:75-87
- 1 2 3 4 5 6 7 8 9 10 11 12 Votteler, J. and Schubert, U. (2008) Human Immunodeficiency Viruses: Molecular Biology. Encyclopedia of Virology. (3rd ed.) 517-525
- ↑ Votteler, J. and Schubert, U. (2008) Human Immunodeficiency Viruses: Molecular Biology. Encyclopedia of Virology (3rd Ed) 517-525
- 1 2 Feinberg, Mark B and Greene, Warner C. (1992) Molecular Insights into human immunodeficiency virus type1 pathogenesis. Current Opinion in Immunology. 4:466-474.
- 1 2 King Steven R (1994). "HIV: Virology and Mechanisms of disease". Annals of Emergency Medicine. 24: 443–449. doi:10.1016/s0196-0644(94)70181-4.
- ↑ Benko, DM; Schwartz, S; Pavlakis, GN; Felber, BK (June 1990). "A novel human immunodeficiency virus type 1 protein, tev, shares sequences with tat, env, and rev proteins.". Journal of Virology. 64 (6): 2505–18. PMID 2186172.
- ↑ Berkhout B (January 1992). "Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis". Nucleic Acids Res. 20 (1): 27–31. doi:10.1093/nar/20.1.27. PMC 310321
 . PMID 1738599.
. PMID 1738599. - ↑ Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R (February 2002). "In vitro evidence for a long range pseudoknot in the 5'-untranslated and matrix coding regions of HIV-1 genomic RNA". J. Biol. Chem. 277 (8): 5995–6004. doi:10.1074/jbc.M108972200. PMID 11744696.
- ↑ Damgaard, CK; Andersen ES; Knudsen B; Gorodkin J; Kjems J (2004). "RNA interactions in the 5' region of the HIV-1 genome". J Mol Biol. 336 (2): 369–379. doi:10.1016/j.jmb.2003.12.010. PMID 14757051.
- ↑ Rong, L; Russell RS; Hu J; Laughrea M; Wainberg MA; Liang C (2003). "Deletion of stem-loop 3 is compensated by second-site mutations within the Gag protein of human immunodeficiency virus type 1". Virology. 314 (1): 221–228. doi:10.1016/S0042-6822(03)00405-7. PMID 14517075.
- ↑ Wang Q, Barr I, Guo F, Lee C (December 2008). "Evidence of a novel RNA secondary structurein the coding region of HIV-1 pol gene". RNA. 14 (12): 2478–88. doi:10.1261/rna.1252608. PMC 2590956
 . PMID 18974280.
. PMID 18974280. - ↑ Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM (2009). "Architecture and Secondary Structure of an Entire HIV-1 RNA Genome". Nature. 460 (7256): 711–6. Bibcode:2009Natur.460..711W. doi:10.1038/nature08237. PMC 2724670
 . PMID 19661910.
. PMID 19661910.
External links
- Hunt R. "HIV and AIDS". Human Immunodeficiency Virus and AIDS. University of South Carolina School of Medicine. Retrieved 2008-08-06.
- Rfam entry for HIV pol-1 stem loop
- 3D model of the complete HIV1 virion
- "3D visualization of HIV virions by cryoelectron tomography". Meth. Enzymol. 483: 267–90. 2010. doi:10.1016/S0076-6879(10)83014-9. PMC 3056484
 . PMID 20888479.
. PMID 20888479.