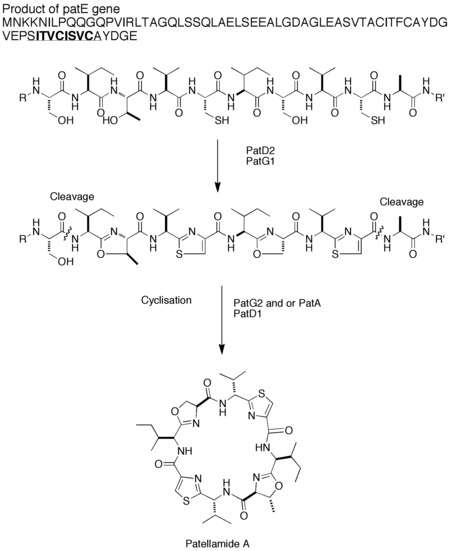
Patellamide A
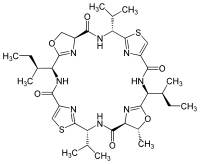 | |
| Identifiers | |
|---|---|
| 81120-73-2 | |
| 3D model (Jmol) | Interactive image |
| ChemSpider | 138571 |
| PubChem | 157454 |
| |
| |
| Properties | |
| C35H50N8O6S2 | |
| Molar mass | 742.96 g·mol−1 |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Patellamide A is a peptide natural product produced by Prochloron didemni, a cyanobacterial symbiont of Lissoclinum patella, and was first isolated in 1981.[1] Patellamide A is one of many didemnid peptides. Other closely related peptides include patellamides B, C, and D and trunkamide. The patellamides and trunkamide show moderate cytotoxicity and activity against multidrug resistant cancer cell lines.[2]
History
Patellamide A was first isolated in 1981 from the tunicate L. patella collected from the reefs of Korror Island, Palau Islands.[1] L. patella has been a rich source of peptide natural products. Aside from the patellamides, the lissoclinamides, ulicyclamide, ulithiacyclamide and ascidiacyclamide were all isolated from L. patella.[3] The absolute stereochemistry was later determined by X-ray crystallography.[4]
Biosynthesis
Patellamide A originates from a ribosomal peptide, making it a member of the RiPP superfamily of natural products. This was determined after genome sequencing of P. didemi showed a lack of non ribosomal peptide synthetases.[2] The biosynthetic gene cluster for patellamide A contains the genes patA, patB, patC, patD, patE, patF and patG.[2] These genes, when introduced into E. coli, cause the production of patellamide A, definitively confirming their responsibility for patellamide A biosynthesis. The gene patE encodes the precursor peptide that contains the primary sequences of patellamides A and C. It has been proposed by Schmidt et al. that this prepatellamide is heterocyclized to form the oxazoline and thiazoline rings by PatD2. It is proposed that PatG1 is subsequently involved in oxidizing the thiazoline rings to the thiazole rings found in patellamide A. The peptide is then cleaved, possibly by PatA or PatG2, and cyclized, the cyclization is likely aided by adenylation by PatD1, forming the two cyclic peptides, patellamides A and C.[2] Although all the amino acids used in the production of patellamide A are L-amino acids, some of the amino acids found in natural patellamide A are the D-epimers. It is proposed that epimerization of these amino acids occurs spontaneously. This was determined by comparison to a similar system, lissoclinamide 7.[5]
References
- 1 2 Ireland, C.; Durso, A.; Newman, R.; Hacker, D. (1982). "Antineoplastic cyclic peptides from the marine tunicate Lissoclinum patella". J. Org. Chem. 47 (10): 1807–1811. doi:10.1021/jo00349a002.
- 1 2 3 4 5 Schmidt, E.; Nelson, J.; Rasko, D.; Sudek, S.; Eisen, J.; Haygood, M.; Ravel, J. (2005). "Patellamide a and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella". Proc. Natl. Acad. Sci. 102 (20): 7315–7320. doi:10.1073/pnas.0501424102. PMC 1091749
 . PMID 15883371.
. PMID 15883371. - ↑ Degnan, B.; Hawkins, C.; Lavin, M.; Mccaffrey, E.; Parry, D.; Vandenbrenk, A.; Watters D. (1989). "New cyclic peptides with cytotoxic activity from the ascidian Lissoclinum patella". J. Med. Chem. 32 (6): 1349–1354. doi:10.1021/jm00126a034. PMID 2724305.
- ↑ In, Y.; Doi, M.; Inoue, M.; Ishida, T. (1994). "Patellamide A, a cytotoxic cyclic peptide from the ascidian Lissoclinum patella". Acta Crystallogr. 50: 432–434. doi:10.1107/s010827019300811x.
- ↑ Milne, B.; Long, P.; Starcevic, A.; Hranueli, D.; Jaspars, M. (2006). "Spontaneity in the patellamide biosynthetic pathway.". Org. Biomol. Chem. 4 (4): 631–638. doi:10.1039/b515938e. PMID 16467937.