Penicillium digitatum
| Penicillium digitatum | |
|---|---|
 | |
| Scientific classification | |
| Kingdom: | Fungi |
| Division: | Ascomycota |
| Subdivision: | Pezizomycotina |
| Class: | Eurotiomycetes |
| Order: | Eurotiales |
| Genus: | Penicillium |
| Species: | P. digitatum |
| Binomial name | |
| Penicillium digitatum (Pers.) Sacc. | |
| Synonyms | |
| |
Penicillium digitatum (/ˌpɛnɪˈsɪlɪəm/digitatum/) is a mesophilic fungus found in the soil of citrus-producing areas.[1][2][3] It is a major source of post-harvest decay in fruits and is responsible for the widespread post-harvest disease in Citrus fruit known as green rot or mould.[1][4] In nature, this necrotrophic wound pathogen grows in filaments and reproduces asexually through the production of conidiophores.[1][5][6] However, P. digitatum can also be cultivated in the laboratory setting.[1] Alongside its pathogenic life cycle, P. digitatum is also involved in other human, animal and plant interactions and is currently being used in the production of immunologically based mycological detection assays for the food industry.[1][7][8]
History and taxonomy
Penicillium digitatum is a species within the Ascomycota division of Fungi. The genus name Penicillium comes from the word "penicillus" which means brush, referring to the branching appearance of the asexual reproductive structures found within this genus.[9] As a species, P. digitatum was first noted as Aspergillus digitatus by Christiaan Hendrik Persoon in 1794 who later adopted the name Monilia digitata in Synopsis methodica fungorum (1801).[10] The synonym M. digitata can also be found in the writings of Elias Magnus Fries in Systema mycologicum (1832).[11] However, the current binomial name comes from the writings of Pier Andrea Saccardo, particularly Fungi italici autographie delineati et colorati (1881).[11]
Growth and morphology
In nature, P. digitatum adopts a filamentous vegetative growth form, producing narrow, septate hyphae.[12] The hyphal cells are haploid, although individual hyphal compartments may contain many genetically identical nuclei.[13] During the reproductive stages of its life cycle, P. digitatum reproduces asexually via the production of asexual spores or conidia.[12] Conidia are borne on a stalk called a conidiophore that can emerge either from a piece of aerial hyphae or from a soil-embedded network of hyphae.[1][12] The conidiophore is usually an asymmetrical, delicate structure with smooth, thin walls.[1][2] Sizes can range from 70-150 µm in length.[1] During development, the conidiophore can branch into three rami to produce a terverticillate structure although biverticillate and other irregular structures are often observed.[1] At the end of each rami, another set of branches called metulae are found. The number of metulae varies with their sizes ranging from 15-30 x 4-6 µm.[2] At the distal end of each metula, conidium-bearing structures called phialides form. Phialides can range in shape from flask-shaped to cylindrical and can be 10-20 µm long.[1] The conidia produced, in turn, are smooth with a shape that can range from spherical to cylindrical although an oval shape is frequently seen.[1][2] They are 6-15 µm long and are produced in chains, with the youngest at the base of each chain.[1][12] Each conidium is haploid and bears only one nucleus.[13] Sexual reproduction in P. digitatum has not been observed.[13]
Penicillium digitatum can also grow on a variety of laboratory media. On Czapek Yeast Extract Agar medium at 25 °C, white colonies grow in a plane, attaining a velvety to deeply floccose texture with colony sizes that are 33-35 mm in diameter.[1] On this medium, olive conidia are produced.[1] The reverse of the plate can be pale or slightly tinted brown.[1] On Malt Extract Agar medium at 25°C, growth is rapid yet rare, forming a velvety surface.[1][2] At first, colonies are yellow-green but ultimately turn olive due to conidial production.[2] Colony diameter can range in size from 35 mm to 70 mm.[1] The reverse of the plate is similar to that observed for Czapek Yeast Extract Agar medium.[1] On 25% Glycerol Nitrate Agar at 25 °C, colony growth is planar yet develops into a think gel with colony size diameter ranging from 6-12 mm.[1] The back of the plate is described as pale or olive.[1] At 5 °C, 25% Glycerol Nitrate Agar supports germination and a colonial growth of up to 3 mm in diameter.[1] This species fails to grow at 37 °C.[1] On Creatine Sucrose Agar at 25 °C, colony size diameter ranges from 4-10 mm.[1] Growth is restricted and medium pH remains around 7.[1] No change on the back of the plate is noted.[1] Growth on media containing orange fruit pieces for seven days at room temperature results in fruit decay accompanied by a characteristic odour.[1] After 14 days at room temperature, the reverse is colourless to light brown.[1]
Ecology
Penicillium digitatum is found in the soil of areas cultivating citrus fruit, predominating in high temperature regions.[1][2] In nature, it is often found alongside the fruits it infects, making species within the Citrus genus its main ecosystem.[1][2] It is only within these species that P. digitatum can complete its life cycle as a necrotroph.[5][13] However, P. digitatum has also been isolated from other food sources.[1] These include hazelnuts, pistachio nuts, kola nuts, black olives, rice, maize and meats.[1] Low levels have also been noted in Southeast Asian peanuts, soybeans and sorghum.[1]
Physiology
Penicillium digitatum is a mesophilic fungus, growing from 6–7 °C (43–45 °F) to a maximum of 37 °C (99 °F), with an optimal growth temperature at 24 °C (75 °F).[1][3] With respect to water activity, P. digitatum has a relatively low tolerance for osmotic stress. The minimum water activity required for growth at 25 °C (77 °F) is 0.90, at 37 °C (99 °F) is 0.95 and at 5 °C (41 °F) is 0.99.[1] Germination does not occur at a water activity of 0.87.[1] In terms of chemicals that influence fungal growth, the minimum growth inhibitory concentration of sorbic acid is 0.02-0.025% at a pH of 4.7 and 0.06-0.08% at a pH of 5.5.[1] Thiamine, on the other hand, has been observed to accelerate fungal growth with the effect being co-metabolically enhanced in the presence of tyrosine, casein or zinc metal.[7] In terms of carbon nutrition, maltose, acetic acid, oxalic acid and tartaric acid support little, if any, growth.[7] However, glucose, fructose, sucrose, galactose, citric acid and malic acid all maintain fungal growth.[7]
Production of ethylene via the Citric acid cycle has been observed in static cultures and is suggested to be connected to mycelial development.[14] Addition of methionine inhibits such cultures but can be utilized for the production of ethylene following a lag phase in shake cultures.[14] The production observed in shake cultures can be inhibited by actinomycin D and cycloheximide and modulated by inorganic phosphate.[14] In addition, aminoethoxyvinyl glycine and methoxyvinyl glycine have been shown to inhibit both shake and static cultures.[14] Production of mycotoxins or secondary metabolites by P. digitatum has not been observed although this species has been shown to be toxic to both shrimp and chicken embryos.[1]
With respect to fungicidal tolerance, there are known strains of P. digitatum resistant to various commonly used fungicides.[1] Reports have been made concerning fungicides thiabendazole, benomyl, imazalil, sodium-o-phenylphenate as well as fungistatic agent, biphenyl, with no prior treatment required in the case of biphenyl.[1][2] The mechanism of P. digitatum resistance to imazalil is suggested to lie in the over-expression of the sterol 14α-demethylase (CYP51) protein caused by a 199 base-pair insertion into the promoter region of the CYP51 gene and/or by duplications of the CYP51 gene.[15]
Human pathogenicity
Species within the Penicillium genus do not generally cause disease in humans.[16] However, being one of the most common producers of indoor moulds, certain species can become pathogenic upon long-term exposure as well as for individuals who are immunocompromised or hyper-sensitized to certain parts of the fungus.[16][17] Spores, proteolytic enzymes and glycoproteins are amongst the components commonly reported as allergens in humans and animal models.[17] Within this context, members of the Penicillium genus have been associated with a variety of immunological manifestations such as Type 1 allergic responses, hypersensitivity pneumonitis (Type 3 responses), and immediate and delayed asthma.[17]
With respect to P. digitatum, this species is known to cause generalized mycosis in humans, although the incidence of such events are very low.[16] Various studies have also noted a presence of circulating antibodies to the extracellular polysaccharide of P. digitatum in both human and rabbit sera.[18] This presence is suggested to be due to the intake of contaminated fruits and/or breathing air contaminated with extracellular polysaccharide.[18] In terms of allergy testing, P. digitatum is present in various clinical allergy test formulations, testing for allergy to moulds.[19] There has been one case report identifying P. digitatum as the cause of a fatal case of pneumonia through molecular methods.[16]
Plant interactions
Post-harvest decays are a main source of fruit loss following harvesting, with the most common source of Citrus fruit decay being infections caused by P. digitatum and P. italicum.[1][4] Penicillium digitatum is responsible for 90% of citrus fruits lost to infection after harvesting and considered the largest cause of post-harvest diseases occurring in Californian citrus fruits.[20] Its widespread impact relates to the post-harvest disease it causes in citrus fruits known as green rot or mould.[6] As a wound pathogen, the disease cycle begins when P. digitatum conidia germinate with release of water and nutrients from the site of injury on the fruit surface.[6][21] After infection at 24°C, rapid growth ensues with active infection taking place within 48 hours and initial symptom onset occurring within 3 days.[3][6] As temperature at time of infection decreases, the delay of initial symptom onset increases.[3] Initial symptoms include a moist depression on the surface which expands as white mycelium colonizes much of its surface.[3] The centre of the mycelial mass eventually turns olive as conidial production begins.[2][3] Near the end of the disease cycle, the fruit eventually decreases in size and develops into an empty, dry shell.[2] This end result is commonly used to distinguish P. digitatum infections from those of P. italicum which produce a blue-green mould and ultimately render the fruit slimy.[2]
Infection with green mould at 25 °C (77 °F) can last 3 to 5 days with the rate of conidial production per infected fruit being as high as 1-2 billion conidia.[21] Annual infections can occur anywhere from December to June and can take place throughout any point during and following harvesting.[6] Transmission can occur mechanically or via conidial dispersal in water or air to fruit surfaces.[3][6] Conidia often reside within soil but can also be found in the air of contaminated storage spaces.[3] Being a wound pathogen, fruit injuries are required for successful fruit infections, with much of these injuries occurring due to improper handling throughout the harvesting process.[3] Injuries can also be caused by other events such as frost and insect bites, and can be as minor as damage to fruit skin oil glands.[3][6] Fallen fruit can also be susceptible to P. digitatum infections as has been noted in Israel, where P. digitatum infects fallen fruit more than P. italicum.[2]
Pathogenicity of P. digitatum is suggested to rely on the acidification of the infected fruit.[1][22] During fruit decay, this species has been observed to make citric acid and gluconic acid and sequester ammonium ions into its cytoplasm.[22]The low pH may aid in the regulation of various gene-encoded pathogenic factors such as polygalactouronases.[1][22] In addition, P. digitatum has also been observed to modify plant defense mechanisms, such as phenylalanine ammonia lyase activity, in the citrus fruits it infects.[22]
Modifications to the disease cycle of P. digitatum have been induced experimentally. For example, P. digitatum has been observed to cause infection in unwounded fruits through mechanical transmission although a higher infection dose was required in such instances.[12] Apples have also been infected to a limited extent.[8] Besides its pathogenic interactions, P. digitatum has also been implicated in naturally accelerating the ripening of green fruits and causing epinastic responses in various plants such as potato, tomato and sunflowers.[7]
Prevention of plant disease
Control of green mould initially relies on the proper handling of fruit before, during and after harvesting.[2][6] Spores can be reduced by removing fallen fruit.[1][6] Risk of injury can be decreased in a variety of ways including, storing fruit in high humidity/ low temperature conditions, and harvesting before irrigation or rainfall in order to minimize fruit susceptibility to peel damage.[6] Degreening practices can also be conducted at humidities above 92% in order to heal injuries.[6]
Chemical control in the form of fungicides is also commonly used.[1] Examples include imazalil, thaibendazole and biphenyl, all of which suppress the reproductive cycle of P. digitatum.[3] Post-harvest chemical treatment usually consists of washes conducted at 40–50 °C (104–122 °F), containing detergents, weak alkalines and fungicides.[1] Californian packinghouses typically use a fungicide cocktail containing sodium o-phenylphenate, imazalil and thiabendazole.[21] In Australia, guazatine is commonly used although this treatment is restricted to the domestic market.[1] In terms of the export market, Generally recognized as safe (GRAS) substances are currently being explored as alternatives.[1] GRAS substances such as sodium bicarbonate, sodium carbonate and ethanol, have displayed an ability to control P. digitatum by decreasing germination rate.[23]
Resistance to common fungicides is currently combated through the use of other chemicals. For example, sodium o-phenylphenate-resistant strains are dealt with via formaldehyde fumigation while imazalil-resistant strains are controlled through the use of pyrimethanil, a fungicide also approved for fighting strains resistant to other fungicides.[1] As fungicide resistance increases globally, other measures of control are being considered including that of biocontrol. Effective biocontrol agents include bacteria such as Bacillus subtilis, Pseudomonas cepacia and Pseudomonas syringae as well as fungi such as Debaryomyces hansenii and Candida guilliermondii.[1] In Clementines and Valencia oranges, Candida oleophila, Pichia anomala and Candida famata have been shown to reduce disease.[1][23] Despite the ability of various biocontrol agents to exhibit antagonistic activity, biocontrol has not been shown to provide complete control over P.digitatum and is therefore commonly used in conjunction with another measure of control.[23] Alternative measures of control include essential oils such as Syzygium aromaticum and Lippia javanica, ultraviolet light, curing, vapour heat and cell-penetrating anti-fungal peptides.[1][24][25]
Laboratory identification
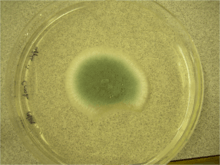
Penicillium digitatum can be identified in the laboratory using a variety of methods. Typically, strains are grown for one week on three chemically defined media under varying temperature conditions.[1] The media used are Czapek Yeast Extract Agar (at 5, 25 and 37 °C), Malt Extract Agar (at 25 °C) and 25% Glycerol Nitrate Agar (at 25°C).[1] The resulting colonial morphology on these media (described in Growth and Morphology above) allows for identification of P. digitatum. Closely related species in the Pencillium genus can be resolved through this approach by using Creatine Sucrose Neutral Agar.[1] Molecular methods can also aid with identification.[1] The genomes of many species belonging to the genus Penicillium remain to be sequenced however, limiting the applicability of such methods.[1] Lastly, P. digitatum can also be distinguished macroscopically by the production of yellow-green to olive conidia and microscopically, by the presence of large philades and conidia.[1]
Industrial uses
Penicillium digitatum is used as a biological tool during the commercial production of latex agglutination kits.[1] Latex agglutination detects Aspergillus and Penicillium species in foods by attaching antibodies specific for the extracellular polysaccharide of P. digitatum to 0.8 µm latex beads.[1] This method has been successful in detecting contamination of grains and processed foods at a limit of detection of 5-10 ng/mL of antigen.[1] In comparison to other detection assays, the latex agglutionation assay exceeds the detection limit of the Enzyme-linked immunosorbent assay (ELISA) and is as effective in detecting Aspergillus and Pencillium species as the ergosterol production assay.[1] However, the latter displays an increased ability to detect Fusarium species when compared to the latex agglutination assay.[1]
External links
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 Pitt, John I.; Hocking, Alisa D. (1985). Fungi and food spoilage (3rd ed). Dordrecht: Springer. ISBN 9780387922072.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Onions, A.H.S. "Penicillium digitatum. C.M.I. Descriptions of Fungi and Bacteria No. 96". Descriptions of Fungi and Bacteria. CAB International Wallingford UK.
- 1 2 3 4 5 6 7 8 9 10 11 Smith, I.M. (1988). European handbook of plant diseases ([Online-Ausg.] ed.). Oxford [Oxfordshire]: Blackwell Scientific Publications. ISBN 0632012226.
- 1 2 Wilson, Charles L.; Wisniewski, Michael E.; Biles, Charles L.; McLaughlin, Randy; Chalutz, Edo; Droby, Samir (1991-06-01). "Biological control of post-harvest diseases of fruits and vegetables: alternatives to synthetic fungicides". Crop Protection. 10 (3): 172–177. doi:10.1016/0261-2194(91)90039-T.
- 1 2 Marcet-Houben, Marina; Ballester, Ana-Rosa; de la Fuente, Beatriz; Harries, Eleonora; Marcos, Jose F.; González-Candelas, Luis; Gabaldón, Toni (2012-01-01). "Genome sequence of the necrotrophic fungus Penicillium digitatum, the main post-harvest pathogen of citrus". BMC Genomics. 13: 646. doi:10.1186/1471-2164-13-646. ISSN 1471-2164. PMC 3532085
 . PMID 23171342.
. PMID 23171342. - 1 2 3 4 5 6 7 8 9 10 11 Brown, G. Eldon. "Citrus Diseases-PostHarvest" (PDF). University of Florida: IFAS Indian River Research and Education Center. University of Florida.
- 1 2 3 4 5 Fergus, Charles L. (1952-03-01). "The Nutrition of Penicillium digitatum Sacc.". Mycologia. 44 (2): 183–199. JSTOR 4547585.
- 1 2 Issues in General Food Research: 2013 Edition. ScholarlyEditions. 2013-05-01. ISBN 9781490106892.
- ↑ Barron, George. "Penicillium italicum and Penicillium digitatum on Orange". George Barron's Website on Fungi.
- ↑ "Penicillium digitatum". Mycobank. CBS-KNAW Fungal Biodiversity Center.
- 1 2 Samson, Robert A; Pitt, John I (2000). Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Amsterdam, The Netherlands: Harwood Acad. Publ. p. 23. ISBN 9058231593.
- 1 2 3 4 5 Peberdy, John F (1987). Penicillium and Acremonium. New York: Plenum Press. ISBN 0306423456.
- 1 2 3 4 Georghiou, G. P. (2012). Pest Resistance to Pesticides. Springer Science & Business Media. ISBN 9781468444667.
- 1 2 3 4 Lieberman, M. (1979-01-01). "Biosynthesis and Action of Ethylene". Annual Review of Plant Physiology. 30 (1): 533–591. doi:10.1146/annurev.pp.30.060179.002533.
- ↑ Price, Claire L; Parker, Josie E; Warrilow, Andrew GS; Kelly, Diane E; Kelly, Steven L (2015-08-01). "Azole fungicides – understanding resistance mechanisms in agricultural fungal pathogens". Pest Management Science. 71 (8): 1054–1058. doi:10.1002/ps.4029. ISSN 1526-4998.
- 1 2 3 4 Oshikata, Chiyako; Tsurikisawa, Naomi; Saito, Akemi; Watanabe, Maiko; Kamata, Yoichi; Tanaka, Maki; Tsuburai, Takahiro; Mitomi, Hiroyuki; Takatori, Kosuke (2013-03-23). "Fatal pneumonia caused by Penicillium digitatum: a case report". BMC Pulmonary Medicine. 13 (1): 16. doi:10.1186/1471-2466-13-16. ISSN 1471-2466. PMC 3614886
 . PMID 23522080.
. PMID 23522080. 
- 1 2 3 Halewyn, Marie-Alix; Chevalier, Pierre. "Penicillium spp.". INSPQ. Institut national de santé publique. Retrieved November 13, 2015.
- 1 2 Notermans, S.; Dufrenne, J.; Wijnands, L. M.; Engel, H. W. B. (1988-01-01). "Human serum antibodies to extracellular polysaccharides (EPS) of moulds". Journal of Medical and Veterinary Mycology. 26 (1): 41–48. doi:10.1080/02681218880000051. ISSN 0268-1218.
- ↑ "Animal Allergens Injection". Drugs.com. Hollister Stier Laboratories LLC. Retrieved November 13, 2015.
- ↑ Ariza, Marta R.; Larsen, Thomas O.; Duus, Jens Ø.; Barrero, Alejandro F. (2002-10-01). "Penicillium digitatum Metabolites on Synthetic Media and Citrus Fruits". Journal of Agricultural and Food Chemistry. 50 (22): 6361–6365. doi:10.1021/jf020398d. ISSN 0021-8561.
- 1 2 3 Holmes, GJ; Eckert, JW (1999). "Sensitivity of Penicillium digitatum and P. italicum to Postharvest Citrus Fungicides in California". Phytopathology. 89: 716–21. doi:10.1094/PHYTO.1999.89.9.716. PMID 18944698.
- 1 2 3 4 Macarisin, D; Cohen, L; Eick, A; Rafael, G; Belausov, E; Wisniewski, M; Droby, S (2007). "Penicillium digitatum Suppresses Production of Hydrogen Peroxide in Host Tissue During Infection of Citrus Fruit". Phytopathology. 97: 1491–500. doi:10.1094/PHYTO-97-11-1491. PMID 18943520.
- 1 2 3 Janisiewicz, Wojciech J.; Korsten, Lise (2002-01-01). "Biological Control of Postharvest Diseases of Fruits". Annual Review of Phytopathology. 40 (1): 411–441. doi:10.1146/annurev.phyto.40.120401.130158. PMID 12147766.
- ↑ Sivakumar, Dharini; Bautista-Baños, Silvia (2014-10-01). "A review on the use of essential oils for postharvest decay control and maintenance of fruit quality during storage". Crop Protection. 64: 27–37. doi:10.1016/j.cropro.2014.05.012.
- ↑ Muñoz, Alberto; Gandía, Mónica; Harries, Eleonora; Carmona, Lourdes; Read, Nick D.; Marcos, Jose F. (2013-01-01). "Understanding the mechanism of action of cell-penetrating antifungal peptides using the rationally designed hexapeptide PAF26 as a model". Fungal Biology Reviews. Revealing diverse modes of action and biological roles of antifungal peptides. 26 (4): 146–155. doi:10.1016/j.fbr.2012.10.003.