Phosphoribosylanthranilate isomerase
| phosphoribosylanthranilate isomerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 3D rendering of Phosophoribosylanthranilate Isomerase | |||||||||
| Identifiers | |||||||||
| EC number | 5.3.1.24 | ||||||||
| CAS number | 37259-82-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
| PRAI | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of mutually generated monomers of dimeric phosphoribosylantranilate isomerase from Thermotoga maritima | |||||||||
| Identifiers | |||||||||
| Symbol | PRAI | ||||||||
| Pfam | PF00697 | ||||||||
| Pfam clan | CL0036 | ||||||||
| InterPro | IPR001240 | ||||||||
| SCOP | 1pii | ||||||||
| SUPERFAMILY | 1pii | ||||||||
| |||||||||
In enzymology, a phosphoribosylanthranilate isomerase (EC 5.3.1.24) is an enzyme that catalyzes the third step of the synthesis of the amino acid tryptophan.[1]
This enzyme participates in the phenylalanine, tyrosine and tryptophan biosynthesis pathway, also known as the aromatic amino acid biosynthesis pathway
In yeast it is encoded by the TRP1 gene.[2]
Reaction
- N-(5-phospho-beta-D-ribosyl)anthranilate 1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate
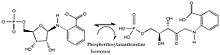
In other words, this enzyme has one substrate, N-(5-phospho-beta-D-ribosyl)anthranilate, and one product, 1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate. As the name phosphoribosylanthranilate isomerase suggests, it functions as an isomerase, rearranging the parts of the molecule without adding or removing molecules or atoms
Nomenclature
This enzyme belongs to the family of isomerases, specifically those intramolecular oxidoreductases interconverting aldoses and ketoses. The systematic name of this enzyme class is N-(5-phospho-beta-D-ribosyl)anthranilate aldose-ketose-isomerase. Other names in common use include:
- PRA isomerase,
- PRAI,
- IGPS:PRAI (indole-3-glycerol-phosphate,
- synthetase/N-5'-phosphoribosylanthranilate isomerase complex), and
- N-(5-phospho-beta-D-ribosyl)anthranilate ketol-isomerase.
Structure
Phosphoribosylanthranilate isomerase (PRAI) is monomeric and labile in most mesophilic microorganisms, but dimeric and stable in the hyperthermophile Thermotoga maritima (tPRAI).[3] The comparison to the known 2.0 A structure of PRAI from Escherichia coli (ePRAI) shows that tPRAI has a TIM-barrel fold, whereas helix alpha5 in ePRAI is replaced by a loop. The subunits of tPRAI associate via the N-terminal faces of their central beta-barrels. Two long, symmetry-related loops that protrude reciprocally into cavities of the other subunit provide for multiple hydrophobic interactions. Moreover, the side chains of the N-terminal methionines and the C-terminal leucines of both subunits are immobilized in a hydrophobic cluster, and the number of salt bridges is increased in tPRAI. These features appear to be mainly responsible for the high thermostability of tPRAI.[4]
Homologous genes
There are homologous genes which produce this enzyme in plant species such as Arabidopsis thaliana and Oryza sativa (Asian Rice). One form of bacterium it is found in Thermotoga maritima.
Phosphoribosylanthranilate isomerase is also found in various forms of fungi such as Kluyveromyces lactis (yeast), Saccharomyces cerevisiae (yeast), and Ashbya gossypii.[5]
References
- ↑ Creighton TE, Yanofsky C (1970). "Chorismate to tryptophan (Escherichia coli) - Anthranilate synthetase, PR transferase, PRA isomerase, InGP synthetase, tryptophan synthetase". Methods Enzymol. 17A: 365–380. doi:10.1016/0076-6879(71)17215-1.
- ↑ "TRP1/YDR007W Summary". Saccharomyces genome database. Stanford University.
- ↑ Thoma R, Hennig M, Sterner R, Kirschner K (March 2000). "Structure and function of mutationally generated monomers of dimeric phosphoribosylanthranilate isomerase from Thermotoga maritima". Structure. 8 (3): 265–76. doi:10.1016/s0969-2126(00)00106-4. PMID 10745009.
- ↑ Hennig M, Sterner R, Kirschner K, Jansonius JN (May 1997). "Crystal structure at 2.0 A resolution of phosphoribosyl anthranilate isomerase from the hyperthermophile Thermotoga maritima: possible determinants of protein stability". Biochemistry. 36 (20): 6009–16. doi:10.1021/bi962718q. PMID 9166771.
- ↑ HomoloGene Database. National Center for Biotechnology Information. Blast search for phosphoribosylanthranilate isomerase. http://www.ncbi.nlm.nih.gov/homologene/?term=phosphoribosylanthranilate%20isomerase
Further reading
This article incorporates text from the public domain Pfam and InterPro IPR001240