Shapiro - Senapathy Algorithm
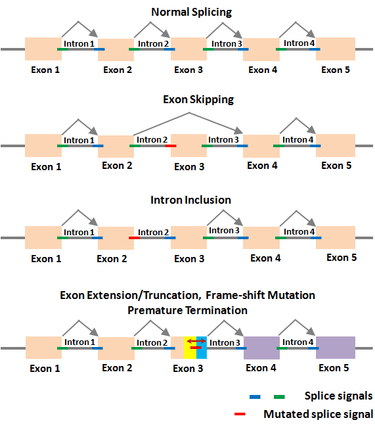
Gene regulation is the main genetic program through which an organism controls its normal functions. Thus, any error in this program caused by mutations will alter the normal state and lead to disease. RNA splicing is increasingly realized to be at the center of gene regulation in eukaryotic organisms, including all animals and plants. In this context, Dr. Periannan Senapathy has pioneered research in the biology of RNA splicing, including understanding of why genes are split, what are splice junction sequences, and why exons are very short and introns are very long.[1][2][3] Based on these findings, he has provided an algorithm (known as Shapiro & Senapathy algorithm, S&S) for predicting the splice sites, exons and genes in animals and plants .[4][5] This algorithm has the ability to discover disease-causing mutations in splice junctions in cancerous and non-cancerous diseases that is being used in major research institutions around the world. The S&S algorithm has been cited in ~3,000 publications on finding splicing mutations in thousands of diseases including many different forms of cancer, non-cancer diseases and plants.
By using the S&S algorithm, mutations and genes that cause different forms of cancer, including, for example, breast cancer [6] ,[7][8] ovarian cancer [9] [10] ,[11] colorectal cancer,[12][13][14] leukemia,[15][16] head and neck cancers,[17][18][19] prostate cancer,[20][21] retinoblastoma,[22][23] squamous cell carcinoma [24] and gastro intestinal cancer [25][26] have been discovered. In addition, other diseases such as diabetes,[27] hypertension,[28][29] marfan syndrome,[30][31][32] cystic fibrosis,[33][34][35] cardiac diseases,[36][37][38] eye disorders,[39] familial hypercholesterolemia,[28] Parkinson disease,[34] have also been uncovered. S&S has also been used in identifying mutations in genes involved in immunodeficiency diseases[40][41][42] and in adverse drug reactions.[43][44] Furthermore, S&S has been implemented for finding splice sites and mutations in several tools such as Human Splice Finder,[45] SROOGLE,[46] Splice-site Analyzer Tool,[47] and dbass (Ensmbl).[48]
It is becoming increasingly known that the pathology in a majority of patients in any disease is caused by mutations in the splicing regions. Thus, applying the S&S technology platform in modern clinical genomics research will advance diagnosis and treatment of human diseases. In addition to its application in thousands of studies involving a variety of diseases, it has been used in finding mutations in drug metabolizing genes that cause adverse reactions. S&S algorithm has also been used in many studies in agricultural plants[49][50][51] and animals.[52][53][54] Using his split gene theory and S&S algorithm, Senapathy's team has developed analytical platforms and several database resources[55][56][57][58] dedicated to the analysis of split genes, splice junctions, and mutations in several genomes including human, animals and plants. Furthermore, based on the statistics of splice signal sequences, Senapathy has developed a functional genome-wide fingerprinting method.[59]
As the mechanism of splicing is inherently complex, the identification of splicing mutations that cause disease is also difficult. The structure of the eukaryotic split genes is highly complex compared to the simple structure of bacterial genes. The reason for this difference is a major question in eukaryotic biology, as it involves the question of how the extremely complex eukaryotic genes could have evolved from the simple genes of prokaryotes. Senapathy has formulated a theory based on his Random-sequence Origin of Split Genes model (ROSG) to explain why the genes of eukaryotes are split into short exon and long intron sequences.[1][2][3] His research has shown that split genes can easily occur within random DNA sequence whereas contiguous genes of bacteria are extremely improbable to occur. These findings show that eukaryotic genes could have originated from prebiotic genetic sequences, and possibly gave rise to eukaryotic genomes. Senapathy has also shown that splice signal sequences that enable the spliceosome to recognize the splicing junctions originated from the stop-codon ends of Open Reading Frames (ORFs) in random sequence.[2]
Studies in evolution of eukaryotic genes and genomes involve the origin of exons, introns and splice junctions, as all eukaryotic genes are split into many exons separated by introns, whereas prokaryotic genes are not. The exons are very short and introns are very long in large genomes such as the human (~3.2 billion bases). Genomes of many invertebrates are also very large such as that of sea urchin (~one billion bases) and contain many introns in their genes. However, the genomes of some animals and plants are relatively small such as those of sea squirt (Ciona Intestinalis – ~115 million bases) and Arabidopsis thaliana (~120 million bases). The genes in the genomes of these organisms are also split into exons and introns, and exhibit basically the same splice junction sequences. The S&S algorithm is being used in researching the evolution of numerous animal and plant genomes.
References
- 1 2 Senapathy, P. (1 April 1986). "Origin of eukaryotic introns: a hypothesis, based on codon distribution statistics in genes, and its implications". Proceedings of the National Academy of Sciences of the United States of America. pp. 2133–2137.
- 1 2 3 Senapathy, P. (1 February 1988). "Possible evolution of splice-junction signals in eukaryotic genes from stop codons". Proceedings of the National Academy of Sciences of the United States of America. pp. 1129–1133.
- 1 2 Senapathy, P. (2 June 1995). "Introns and the origin of protein-coding genes". Science. 268 (5215): 1366–1367. doi:10.1126/science.7761858. ISSN 0036-8075.
- ↑ Shapiro, M. B.; Senapathy, P. (11 September 1987). "RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression". Nucleic Acids Research. pp. 7155–7174.
- ↑ Senapathy, P.; Shapiro, M. B.; Harris, N. L. (1 January 1990). "Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project". Methods in Enzymology. pp. 252–278.
- ↑ Damiola, Francesca; Schultz, Inès; Barjhoux, Laure; Sornin, Valérie; Dondon, Marie-Gabrielle; Eon-Marchais, Séverine; Marcou, Morgane; Investigators, The GENESIS Study; Caron, Olivier; Gauthier-Villars, Marion; Pauw, Antoine de; Luporsi, Elisabeth; Berthet, Pascaline; Delnatte, Capucine; Bonadona, Valérie; Maugard, Christine; Pujol, Pascal; Lasset, Christine; Longy, Michel; Bignon, Yves-Jean; Fricker, Jean-Pierre; Andrieu, Nadine; Sinilnikova, Olga M.; Stoppa-Lyonnet, Dominique; Mazoyer, Sylvie; Muller, Danièle (12 November 2015). "Mutation analysis of PALB2 gene in French breast cancer families". Breast Cancer Research and Treatment. pp. 463–471. doi:10.1007/s10549-015-3625-7.
- ↑ "SciELO - Scientific electronic library online". www.scielo.cl.
- ↑ Di Giacomo, Daniela; Gaildrat, Pascaline; Abuli, Anna; Abdat, Julie; Frébourg, Thierry; Tosi, Mario; Martins, Alexandra (1 November 2013). "Functional Analysis of a Large set of BRCA2 exon 7 Variants Highlights the Predictive Value of Hexamer Scores in Detecting Alterations of Exonic Splicing Regulatory Elements". Human Mutation. 34 (11): 1547–1557. doi:10.1002/humu.22428. ISSN 1098-1004.
- ↑ Kato, Chise; Fujii, Kentaro; Arai, Yuto; Hatsuse, Hiromi; Nagao, Kazuaki; Takayama, Yoshinaga; Kameyama, Kouzou; Fujii, Katsunori; Miyashita, Toshiyuki (25 August 2016). "Nevoid basal cell carcinoma syndrome caused by splicing mutations in the PTCH1 gene". Familial Cancer: 1–8. doi:10.1007/s10689-016-9924-2. ISSN 1389-9600.
- ↑ Kreimann, Erica; Ratajska, Magdalena; Kuzniacka, Alina; Demacopulo, Brenda; Stukan, Maciej; Limon, Janusz (1 December 2015). "A novel splicing mutation in the SLC9A3R1 gene in tumors from ovarian cancer patients". Oncology Letters. 10 (6). doi:10.3892/ol.2015.3796. ISSN 1792-1074.
- ↑ Welander, Jenny; Larsson, Catharina; Bäckdahl, Martin; Hareni, Niyaz; Sivlér, Tobias; Brauckhoff, Michael; Söderkvist, Peter; Gimm, Oliver (15 December 2012). "Integrative genomics reveals frequent somatic NF1 mutations in sporadic pheochromocytomas". Human Molecular Genetics. 21 (26): 5406–5416. doi:10.1093/hmg/dds402. ISSN 0964-6906.
- ↑ Mensenkamp, Arjen R.; Vogelaar, Ingrid P.; van Zelst–Stams, Wendy A. G.; Goossens, Monique; Ouchene, Hicham; Hendriks–Cornelissen, Sandra J. B.; Kwint, Michael P.; Hoogerbrugge, Nicoline; Nagtegaal, Iris D.; Ligtenberg, Marjolijn J. L. (1 March 2014). "Somatic Mutations in MLH1 and MSH2 Are a Frequent Cause of Mismatch-Repair Deficiency in Lynch Syndrome-Like Tumors". Gastroenterology. 146 (3): 643–646.e8. doi:10.1053/j.gastro.2013.12.002.
- ↑ "Germline MLH1 Mutations Are Frequently Identified in Lynch S... : The American Journal of Surgical Pathology". LWW. doi:10.1097/PAS.0000000000000425.
- ↑ Degrolard-Courcet, Emilie; Sokolowska, Joanna; Padeano, Marie-Martine; Guiu, Séverine; Bronner, Myriam; Chery, Carole; Coron, Fanny; Lepage, Côme; Chapusot, Caroline; Loustalot, Catherine; Jouve, Jean-Louis; Hatem, Cyril; Ferrant, Emmanuelle; Martin, Laurent; Coutant, Charles; Baurand, Amandine; Couillault, Gérard; Delignette, Alexandra; El Chehadeh, Salima; Lizard, Sarab; Arnould, Laurent; Fumoleau, Pierre; Callier, Patrick; Mugneret, Francine; Philippe, Christophe; Frebourg, Thierry; Jonveaux, Philippe; Faivre, Laurence (1 August 2014). "Development of primary early-onset colorectal cancers due to biallelic mutations of the FANCD1/BRCA2 gene". European Journal of Human Genetics. 22 (8): 979–987. doi:10.1038/ejhg.2013.278. ISSN 1018-4813.
- ↑ Eggington, J.m.; Bowles, K.r.; Moyes, K.; Manley, S.; Esterling, L.; Sizemore, S.; Rosenthal, E.; Theisen, A.; Saam, J.; Arnell, C.; Pruss, D.; Bennett, J.; Burbidge, L.a.; Roa, B.; Wenstrup, R.j. (1 September 2014). "A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes". Clinical Genetics. 86 (3): 229–237. doi:10.1111/cge.12315. ISSN 1399-0004.
- ↑ Toki, Tsutomu; Kanezaki, Rika; Kobayashi, Eri; Kaneko, Hiroshi; Suzuki, Mikiko; Wang, Runan; Terui, Kiminori; Kanegane, Hirokazu; Maeda, Miho; Endo, Mikiya; Mizuochi, Tatsuki; Adachi, Souichi; Hayashi, Yasuhide; Yamamoto, Masayuki; Shimizu, Ritsuko; Ito, Etsuro (18 April 2013). "Naturally occurring oncogenic GATA1 mutants with internal deletions in transient abnormal myelopoiesis in Down syndrome". Blood. pp. 3181–3184. doi:10.1182/blood-2012-01-405746.
- ↑ van Kuilenburg, André B. P.; Meijer, Judith; Mul, Adri N. P. M.; Meinsma, Rutger; Schmid, Veronika; Dobritzsch, Doreen; Hennekam, Raoul C. M.; Mannens, Marcel M. A. M.; Kiechle, Marion; Etienne-Grimaldi, Marie-Christine; Klümpen, Heinz-Josef; Maring, Jan Gerard; Derleyn, Veerle A.; Maartense, Ed; Milano, Gérard; Vijzelaar, Raymon; Gross, Eva (25 November 2016). "Intragenic deletions and a deep intronic mutation affecting pre-mRNA splicing in the dihydropyrimidine dehydrogenase gene as novel mechanisms causing 5-fluorouracil toxicity". Human Genetics. pp. 529–538. doi:10.1007/s00439-010-0879-3.
- ↑ Hildebrand, Michael S; Tankard, Rick; Gazina, Elena V; Damiano, John A; Lawrence, Kate M; Dahl, Hans-Henrik M; Regan, Brigid M; Shearer, Aiden Eliot; Smith, Richard J H; Marini, Carla; Guerrini, Renzo; Labate, Angelo; Gambardella, Antonio; Tinuper, Paolo; Lichetta, Laura; Baldassari, Sara; Bisulli, Francesca; Pippucci, Tommaso; Scheffer, Ingrid E; Reid, Christopher A; Petrou, Steven; Bahlo, Melanie; Berkovic, Samuel F (25 November 2016). "PRIMA1 mutation: a new cause of nocturnal frontal lobe epilepsy". Annals of Clinical and Translational Neurology. pp. 821–830. doi:10.1002/acn3.224.
- ↑ Hildebrand, Michael S.; Tankard, Rick; Gazina, Elena V.; Damiano, John A.; Lawrence, Kate M.; Dahl, Hans-Henrik M.; Regan, Brigid M.; Shearer, Aiden Eliot; Smith, Richard J. H.; Marini, Carla; Guerrini, Renzo; Labate, Angelo; Gambardella, Antonio; Tinuper, Paolo; Lichetta, Laura; Baldassari, Sara; Bisulli, Francesca; Pippucci, Tommaso; Scheffer, Ingrid E.; Reid, Christopher A.; Petrou, Steven; Bahlo, Melanie; Berkovic, Samuel F. (1 August 2015). "PRIMA1 mutation: a new cause of nocturnal frontal lobe epilepsy". Annals of Clinical and Translational Neurology. 2 (8): 821–830. doi:10.1002/acn3.224. ISSN 2328-9503.
- ↑ Nishida, Atsushi; Minegishi, Maki; Takeuchi, Atsuko; Niba, Emma Tabe Eko; Awano, Hiroyuki; Lee, Tomoko; Iijima, Kazumoto; Takeshima, Yasuhiro; Matsuo, Masafumi (1 June 2015). "Tissue- and case-specific retention of intron 40 in mature dystrophin mRNA". Journal of Human Genetics. pp. 327–333. doi:10.1038/jhg.2015.24.
- ↑ Wittler, Lars; Hilger, Alina; Proske, Judith; Pennimpede, Tracie; Draaken, Markus; Ebert, Anne-Karoline; Rösch, Wolfgang; Stein, Raimund; Nöthen, Markus M.; Reutter, Heiko; Ludwig, Michael (15 September 2012). "Murine expression and mutation analyses of the prostate androgen-regulated mucin-like protein 1 (Parm1) gene, a candidate for human epispadias". Gene. 506 (2): 392–395. doi:10.1016/j.gene.2012.06.082.
- ↑ Hung, Chia-Cheng; Lin, Shin-Yu; Lee, Chien-Nan; Chen, Chih-Ping; Lin, Shuan-Pei; Chao, Mei-Chyn; Chiou, Shyh-Shin; Su, Yi-Ning (1 January 2011). "Low penetrance of retinoblastoma for p.V654L mutation of the RB1 gene". BMC Medical Genetics. 12: 76. doi:10.1186/1471-2350-12-76. ISSN 1471-2350.
- ↑ Zhang, Katherine; Nowak, Inga; Rushlow, Diane; Gallie, Brenda L.; Lohmann, Dietmar R. (1 April 2008). "Patterns of missplicing caused by RB1 gene mutations in patients with retinoblastoma and association with phenotypic expression". Human Mutation. 29 (4): 475–484. doi:10.1002/humu.20664. ISSN 1098-1004.
- ↑ Morrison, Arianne; Chekaluk, Yvonne; Bacares, Ruben; Ladanyi, Marc; Zhang, Liying (1 April 2015). "BAP1 Missense Mutation c.2054 A>T (p.E685V) Completely Disrupts Normal Splicing through Creation of a Novel 5' Splice Site in a Human Mesothelioma Cell Line". PLOS ONE. 10 (4): e0119224. doi:10.1371/journal.pone.0119224. ISSN 1932-6203.
- ↑ Wang, Ya‑Ping (1 May 2013). "Missense mutations of MLH1 and MSH2 genes detected in patients with gastrointestinal cancer are associated with exonic splicing enhancers and silencers". Oncology Letters. 5 (5). doi:10.3892/ol.2013.1243. ISSN 1792-1074.
- ↑ van der Post, Rachel S.; Vogelaar, Ingrid P.; Manders, Peggy; van der Kolk, Lizet E.; Cats, Annemieke; van Hest, Liselotte P.; Sijmons, Rolf; Aalfs, Cora M.; Ausems, Margreet G. E. M.; Gómez García, Encarna B.; Wagner, Anja; Hes, Frederik J.; Arts, Neeltje; Mensenkamp, Arjen R.; van Krieken, J. Han; Hoogerbrugge, Nicoline; Ligtenberg, Marjolijn J. L. (1 October 2015). "Accuracy of Hereditary Diffuse Gastric Cancer Testing Criteria and Outcomes in Patients With a Germline Mutation in CDH1". Gastroenterology. 149 (4): 897–906.e19. doi:10.1053/j.gastro.2015.06.003.
- ↑ Danzig, Jennifer; Levine, Michael A. (1 October 2012). "A novel intronic mutation in SHOX causes short stature by disrupting a splice acceptor site: direct demonstration of aberrant splicing by expression of a minigene in HEK-293T cells". Journal of Pediatric Endocrinology and Metabolism. 25 (9-10). doi:10.1515/jpem-2012-0173. ISSN 2191-0251.
- 1 2 Al-Khateeb, Alyaa; Zahri, Mohd K.; Mohamed, Mohd S.; Sasongko, Teguh H.; Ibrahim, Suhairi; Yusof, Zurkurnai; Zilfalil, Bin A. (1 January 2011). "Analysis of sequence variations in low-density lipoprotein receptor gene among Malaysian patients with familial hypercholesterolemia". BMC Medical Genetics. 12: 40. doi:10.1186/1471-2350-12-40. ISSN 1471-2350.
- ↑ Vilarinho, Laura; Tafulo, Sandra; Sibilio, Michelina; Kok, Fernando; Fontana, Federica; Diogo, Luisa; Venâncio, Margarida; Ferreira, Mariana; Nogueira, Celia; Valongo, Carla; Parenti, Giancarlo; Amorim, António; Azevedo, Luisa (13 November 2009). "Identification of novel L2HGDH gene mutations and update of the pathological spectrum". Journal of Human Genetics. 55 (1): 55–58. doi:10.1038/jhg.2009.110. ISSN 1434-5161.
- ↑ Evangelisti, L.; Lucarini, L.; Attanasio, M.; Lapini, I.; Giusti, B.; Porciani, C.; Gensini, G. F.; Abbate, R.; Pepe, G. (1 September 2010). "A single heterozygous nucleotide substitution displays two different altered mechanisms in the FBN1 gene of five Italian marfan patients". European Journal of Medical Genetics. 53 (5): 299–302. doi:10.1016/j.ejmg.2010.06.002.
- ↑ Dietz, Harry C.; McIntosh, Iain; Sakai, Lynn Y.; Corson, Glen M.; Chalberg, Stephen C.; Pyeritz, Reed E.; Francomano, Clair A. (1 August 1993). "Four Novel FBN1 Mutations: Significance for Mutant Transcript Level and EGF-like Domain Calcium Binding in the Pathogenesis of Marfan Syndrome". Genomics. 17 (2): 468–475. doi:10.1006/geno.1993.1349.
- ↑ Attanasio, M.; Lapini, I.; Evangelisti, L.; Lucarini, L.; Giusti, B.; Porciani, Mc; Fattori, R.; Anichini, C.; Abbate, R.; Gensini, Gf; Pepe, G. (1 July 2008). "FBN1 mutation screening of patients with Marfan syndrome and related disorders: detection of 46 novel FBN1 mutations". Clinical Genetics. pp. 39–46. doi:10.1111/j.1399-0004.2008.01007.x.
- ↑ Palhais, Bruno; Præstegaard, Veronica S.; Sabaratnam, Rugivan; Doktor, Thomas Koed; Lutz, Seraina; Burda, Patricie; Suormala, Terttu; Baumgartner, Matthias; Fowler, Brian; Bruun, Gitte Hoffmann; Andersen, Henriette Skovgaard; Kožich, Viktor; Andresen, Brage Storstein (19 May 2015). "Splice-shifting oligonucleotide (SSO) mediated blocking of an exonic splicing enhancer (ESE) created by the prevalent c.903+469T>C MTRR mutation corrects splicing and restores enzyme activity in patient cells". Nucleic Acids Research. 43 (9): 4627–4639. doi:10.1093/nar/gkv275. ISSN 0305-1048.
- 1 2 Lu, Chin-Song; Lai, Szu-Chia; Wu, Ruey-Meei; Weng, Yi-Hsin; Huang, Chia-Ling; Chen, Rou-Shayn; Chang, Hsiu-Chen; Wu-Chou, Yah-Huei; Yeh, Tu-Hsueh (1 March 2012). "PLA2G6 mutations in PARK14-linked young-onset parkinsonism and sporadic Parkinson's disease". American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 159B (2): 183–191. doi:10.1002/ajmg.b.32012. ISSN 1552-485X.
- ↑ Niroula, Abhishek; Vihinen, Mauno (1 June 2016). "Variation Interpretation Predictors: Principles, Types, Performance, and Choice". Human Mutation. pp. 579–597. doi:10.1002/humu.22987.
- ↑ Rani, Deepa Selvi; Nallari, Pratibha; Priyamvada, Singh; Narasimhan, Calambur; Singh, Lalji; Thangaraj, Kumarasamy (1 January 2012). "High prevalence of Arginine to Glutamine Substitution at 98, 141 and 162 positions in Troponin I (TNNI3) associated with hypertrophic cardiomyopathy among Indians". BMC Medical Genetics. 13: 69. doi:10.1186/1471-2350-13-69. ISSN 1471-2350.
- ↑ Goldstein, Jennifer L.; Austin, Stephanie L.; Boyette, Keri; Kanaly, Angela; Veerapandiyan, Aravind; Rehder, Catherine; Kishnani, Priya S.; Bali, Deeksha S. (1 July 2010). "Molecular analysis of the AGL gene: Identification of 25 novel mutations and evidence of genetic heterogeneity in patients with Glycogen Storage Disease Type III". Genetics in Medicine. 12 (7): 424–430. doi:10.1097/GIM.0b013e3181d94eaa. ISSN 1098-3600.
- ↑ Juan-Mateu, Jonàs; González-Quereda, Lidia; Rodríguez, Maria José; Verdura, Edgard; Lázaro, Kira; Jou, Cristina; Nascimento, Andrés; Jiménez-Mallebrera, Cecilia; Colomer, Jaume; Monges, Soledad; Lubieniecki, Fabiana; Foncuberta, Maria Eugenia; Pascual-Pascual, Samuel Ignacio; Molano, Jesús; Baiget, Montserrat; Gallano, Pia (25 March 2013). "Interplay between DMD Point Mutations and Splicing Signals in Dystrophinopathy Phenotypes". PLOS ONE. 8 (3): e59916. doi:10.1371/journal.pone.0059916. ISSN 1932-6203.
- ↑ Littink, Karin W.; Koenekoop, Robert K.; Born, L. Ingeborgh van den; Collin, Rob W. J.; Moruz, Luminita; Veltman, Joris A.; Roosing, Susanne; Zonneveld, Marijke N.; Omar, Amer; Darvish, Mahshad; Lopez, Irma; Kroes, Hester Y.; Genderen, Maria M. van; Hoyng, Carel B.; Rohrschneider, Klaus; Schooneveld, Mary J. van; Cremers, Frans P. M.; Hollander, Anneke I. den (1 November 2010). "Homozygosity Mapping in Patients with Cone–Rod Dystrophy: Novel Mutations and Clinical Characterizations". Investigative Opthalmology & Visual Science. 51 (11). doi:10.1167/iovs.10-5797. ISSN 1552-5783.
- ↑ Santisteban, I; Arredondo-Vega, F X; Kelly, S; Mary, A; Fischer, A; Hummell, D S; Lawton, A; Sorensen, R U; Stiehm, E R; Uribe, L (1 November 1993). "Novel splicing, missense, and deletion mutations in seven adenosine deaminase-deficient patients with late/delayed onset of combined immunodeficiency disease. Contribution of genotype to phenotype.". Journal of Clinical Investigation. 92 (5): 2291–2302. doi:10.1172/JCI116833. ISSN 0021-9738.
- ↑ Coffey, Alison J.; Brooksbank, Robert A.; Brandau, Oliver; Oohashi, Toshitaka; Howell, Gareth R.; Bye, Jacqueline M.; Cahn, Anthony P.; Durham, Jillian; Heath, Paul; Wray, Paul; Pavitt, Rebecca; Wilkinson, Jane; Leversha, Margaret; Huckle, Elizabeth; Shaw-Smith, Charles J.; Dunham, Andrew; Rhodes, Susan; Schuster, Volker; Porta, Giovanni; Yin, Luo; Serafini, Paola; Sylla, Bakary; Zollo, Massimo; Franco, Brunella; Bolino, Alessandra; Seri, Marco; Lanyi, Arpad; Davis, Jack R.; Webster, David; Harris, Ann; Lenoir, Gilbert; de St Basile, Genevieve; Jones, Alison; Behloradsky, Bernd H.; Achatz, Helene; Murken, Jan; Fassler, Reinhard; Sumegi, Janos; Romeo, Giovanni; Vaudin, Mark; Ross, Mark T.; Meindl, Alfons; Bentley, David R. (1 October 1998). "Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene". Nature Genetics. 20 (2): 129–135. doi:10.1038/2424. ISSN 1061-4036.
- ↑ Mueller, Nancy; van Bel, Nikki; Berkhout, Ben; Das, Atze T. (1 November 2014). "HIV-1 splicing at the major splice donor site is restricted by RNA structure". Virology. 468–470: 609–620. doi:10.1016/j.virol.2014.09.018.
- ↑ Vreken, P.; Kuilenburg, A. B. P. Van; Meinsma, R.; Smit, G. P. A.; Bakker, H. D.; Abreu, R. A. De; Gennip, A. H. van. "A point mutation in an invariant splice donor site leads to exon skipping in two unrelated Dutch patients with dihydropyrimidine dehydrogenase deficiency". Journal of Inherited Metabolic Disease. pp. 645–654. doi:10.1007/BF01799841.
- ↑ Kuilenburg, André B. P. van; Meijer, Judith; Mul, Adri N. P. M.; Meinsma, Rutger; Schmid, Veronika; Dobritzsch, Doreen; Hennekam, Raoul C. M.; Mannens, Marcel M. A. M.; Kiechle, Marion; Etienne-Grimaldi, Marie-Christine; Klümpen, Heinz-Josef; Maring, Jan Gerard; Derleyn, Veerle A.; Maartense, Ed; Milano, Gérard; Vijzelaar, Raymon; Gross, Eva (29 August 2010). "Intragenic deletions and a deep intronic mutation affecting pre-mRNA splicing in the dihydropyrimidine dehydrogenase gene as novel mechanisms causing 5-fluorouracil toxicity". Human Genetics. pp. 529–538. doi:10.1007/s00439-010-0879-3.
- ↑ Desmet, François-Olivier; Hamroun, Dalil; Lalande, Marine; Collod-Béroud, Gwenaëlle; Claustres, Mireille; Béroud, Christophe (1 May 2009). "Human Splicing Finder: an online bioinformatics tool to predict splicing signals". Nucleic Acids Research. pp. e67. doi:10.1093/nar/gkp215.
- ↑ Schwartz, Schraga; Hall, Eitan; Ast, Gil (1 July 2009). "SROOGLE: webserver for integrative, user-friendly visualization of splicing signals". Nucleic Acids Research. pp. W189–W192. doi:10.1093/nar/gkp320.
- ↑ "Splice-Site Analyzer Tool". ibis.tau.ac.il.
- ↑ Southampton, University of. "DBASS3 - Aberrant Splice Site Database". www.dbass.org.uk.
- ↑ Philip, Anna; Syamaladevi, Divya P.; Chakravarthi, M.; Gopinath, K.; Subramonian, N. (19 March 2013). "5′ Regulatory region of ubiquitin 2 gene from Porteresia coarctata makes efficient promoters for transgene expression in monocots and dicots". Plant Cell Reports. 32 (8): 1199–1210. doi:10.1007/s00299-013-1416-3. ISSN 0721-7714.
- ↑ Giammaria, Verónica; Grandellis, Carolina; Bachmann, Sandra; Gargantini, Pablo Rubén; Feingold, Sergio Enrique; Bryan, Glenn; Ulloa, Rita María (4 December 2010). "StCDPK2 expression and activity reveal a highly responsive potato calcium-dependent protein kinase involved in light signalling". Planta. 233 (3): 593–609. doi:10.1007/s00425-010-1319-2. ISSN 0032-0935.
- ↑ Sun, Youwei; He, Zhonghu; Ma, Wujun; Xia, Xianchun (3 November 2010). "Alternative splicing in the coding region of Ppo-A1 directly influences the polyphenol oxidase activity in common wheat (Triticum aestivum L.)". Functional & Integrative Genomics. 11 (1): 85–93. doi:10.1007/s10142-010-0201-4. ISSN 1438-793X.
- ↑ Koo, Taeyoung; Popplewell, Linda; Athanasopoulos, Takis; Dickson, George (5 November 2013). "Triple Trans-Splicing Adeno-Associated Virus Vectors Capable of Transferring the Coding Sequence for Full-Length Dystrophin Protein into Dystrophic Mice". Human Gene Therapy. pp. 98–108. doi:10.1089/hum.2013.164.
- ↑ Meher, Prabina Kumar; Sahu, Tanmaya Kumar; Rao, A. R.; Wahi, S. D. (1 January 2016). "Identification of donor splice sites using support vector machine: a computational approach based on positional, compositional and dependency features". Algorithms for Molecular Biology. 11: 16. doi:10.1186/s13015-016-0078-4. ISSN 1748-7188.
- ↑ Itoh, Hitomi; Washio, Takanori; Tomita, Masaru (1 July 2004). "Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes". RNA. 10 (7): 1005–1018. doi:10.1261/rna.5221604. ISSN 1355-8382.
- ↑ Bhasi, Ashwini; Philip, Philge; Manikandan, Vinu; Senapathy, Periannan (1 January 2009). "ExDom: an integrated database for comparative analysis of the exon–intron structures of protein domains in eukaryotes". Nucleic Acids Research. 37 (suppl 1): D703–D711. doi:10.1093/nar/gkn746. ISSN 0305-1048.
- ↑ Bhasi, Ashwini; Philip, Philge; Sreedharan, Vipin T.; Senapathy, Periannan (1 July 2009). "AspAlt: A tool for inter-database, inter-genomic and user-specific comparative analysis of alternative transcription and alternative splicing in 46 eukaryotes". Genomics. pp. 48–54. doi:10.1016/j.ygeno.2009.02.006.
- ↑ Bhasi, Ashwini; Pandey, Ram Vinay; Utharasamy, Suriya Prabha; Senapathy, Periannan (15 July 2007). "EuSplice: a unified resource for the analysis of splice signals and alternative splicing in eukaryotic genes". Bioinformatics (Oxford, England). pp. 1815–1823. doi:10.1093/bioinformatics/btm084.
- ↑ Bhasi, Ashwini; Senalik, Doug; Simon, Philipp W; Kumar, Brajendra; Manikandan, Vinu; Philip, Philge; Senapathy, Periannan (6 August 2010). "RoBuST: an integrated genomics resource for the root and bulb crop families Apiaceae and Alliaceae". BMC Plant Biology. 10: 161. doi:10.1186/1471-2229-10-161. ISSN 1471-2229.
- ↑ Senapathy, Periannan; Bhasi, Ashwini; Mattox, Jeffrey; Dhandapany, Perundurai S.; Sadayappan, Sakthivel (16 June 2010). "Targeted Genome-Wide Enrichment of Functional Regions". PLOS ONE. 5 (6): e11138. doi:10.1371/journal.pone.0011138. ISSN 1932-6203.