XRCC3
| View/Edit Human | View/Edit Mouse |
DNA repair protein XRCC3 is a protein that in humans is encoded by the XRCC3 gene.[3]
Function
This gene encodes a member of the RecA/Rad51-related protein family that participates in homologous recombination to maintain chromosome stability and repair DNA damage. This gene functionally complements Chinese hamster irs1SF, a repair-deficient mutant that exhibits hypersensitivity to a number of different DNA-damaging agents and is chromosomally unstable. A rare microsatellite polymorphism in this gene is associated with cancer in patients of varying radiosensitivity.[4]
The XRCC3 protein is one of five paralogs of RAD51, including RAD51B (RAD51L1), RAD51C (RAD51L2), RAD51D (RAD51L3), XRCC2 and XRCC3. They each share about 25% amino acid sequence identity with RAD51 and each other.[5]
The RAD51 paralogs are all required for efficient DNA double-strand break repair by homologous recombination and depletion of any paralog results in significant decreases in homologous recombination frequency.[6]
Two paralogs form a complex designated CX3 (RAD51C-XRCC3). Four paralogs form a second complex designated BCDX2 (RAD51B-RAD51C-RAD51D-XRCC2). These two complexes act at two different stages of homologous recombinational DNA repair.
The CX3 complex acts downstream of RAD51, after its recruitment to damage sites.[6] The CX3 complex associates with Holliday junction resolvase activity, probably in a role of stabilizing gene conversion tracts.[6]
The BCDX2 complex is responsible for RAD51 recruitment or stabilization at damage sites.[6] The BCDX2 complex appears to act by facilitating the assembly or stability of the RAD51 nucleoprotein filament.
Interactions
XRCC3 has been shown to interact with RAD51C.[7][8][9][10]
Epigenetic deficiency in cancer
There is an epigenetic cause of XRCC3 deficiency that appears to increase cancer risk. This is the repression of XRCC3 by over-expression of EZH2 protein.
Increased expression of EZH2 leads to epigenetic repression of RAD51 paralogs, including XRCC3, and thus reduces homologous recombinational repair.[11] This reduction was proposed to be a cause of breast cancer.[11] EZH2 is the catalytic subunit of Polycomb Repressor Complex 2 (PRC2) which catalyzes methylation of histone H3 at lysine 27 (H3K27me) and mediates gene silencing of target genes via local chromatin reorganization.[12] EZH2 protein is up-regulated in numerous cancers.[12][13] EZH2 mRNA is up-regulated, on average, 7.5-fold in breast cancer, and between 40% to 75% of breast cancers have over-expressed EZH2 protein.[14]
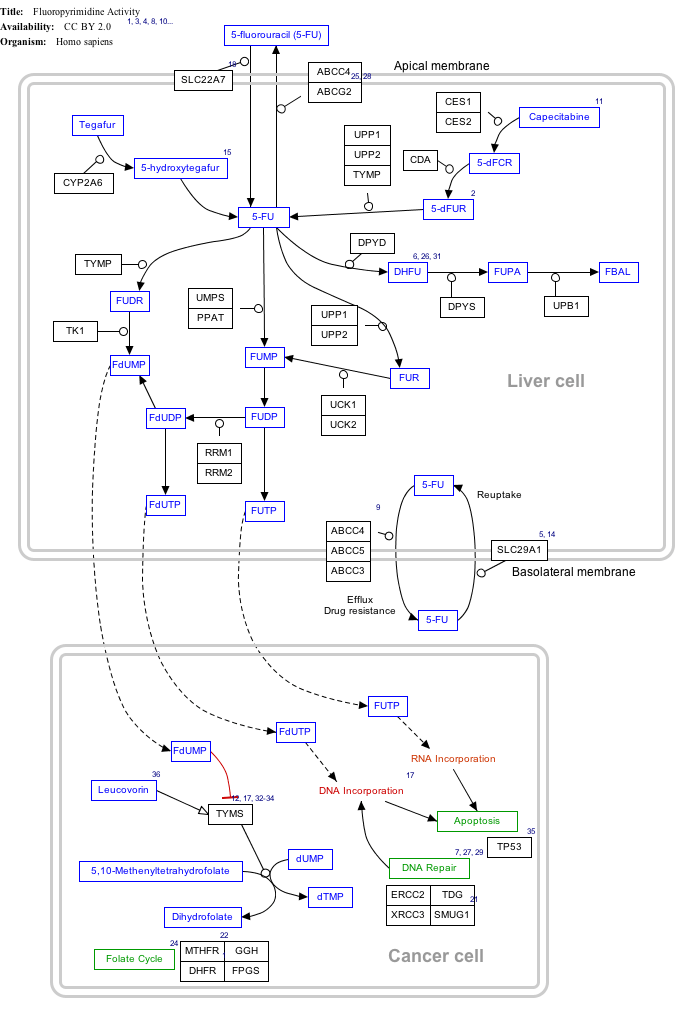
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
Fluorouracil (5-FU) Activity edit
- ↑ The interactive pathway map can be edited at WikiPathways: "FluoropyrimidineActivity_WP1601".
See also
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, Liu N, Legerski RJ, Thompson LH (August 1995). "Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene". Proc Natl Acad Sci U S A. 92 (14): 6354–6358. doi:10.1073/pnas.92.14.6354. PMC 41516
 . PMID 7603995.
. PMID 7603995. - ↑ "Entrez Gene: XRCC3 X-ray repair complementing defective repair in Chinese hamster cells 3".
- ↑ Miller KA, Sawicka D, Barsky D, Albala JS (2004). "Domain mapping of the Rad51 paralog protein complexes". Nucleic Acids Res. 32 (1): 169–78. doi:10.1093/nar/gkg925. PMC 373258
 . PMID 14704354.
. PMID 14704354. - 1 2 3 4 Chun J, Buechelmaier ES, Powell SN (2013). "Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway". Mol. Cell. Biol. 33 (2): 387–95. doi:10.1128/MCB.00465-12. PMC 3554112
 . PMID 23149936.
. PMID 23149936. - ↑ Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, Ananth S, Davies A, Masson JY, Moses R, West SC, de Winter JP, Ashworth A, Jones NJ, Mathew CG (June 2004). "Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways". Hum. Mol. Genet. 13 (12): 1241–8. doi:10.1093/hmg/ddh135. PMID 15115758.
- ↑ Miller KA, Yoshikawa DM, McConnell IR, Clark R, Schild D, Albala JS (March 2002). "RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51". J. Biol. Chem. 277 (10): 8406–11. doi:10.1074/jbc.M108306200. PMID 11744692.
- ↑ Liu N, Schild D, Thelen MP, Thompson LH (February 2002). "Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells". Nucleic Acids Res. 30 (4): 1009–15. doi:10.1093/nar/30.4.1009. PMC 100342
 . PMID 11842113.
. PMID 11842113. - ↑ Kurumizaka H, Ikawa S, Nakada M, Eda K, Kagawa W, Takata M, Takeda S, Yokoyama S, Shibata T (May 2001). "Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C". Proc. Natl. Acad. Sci. U.S.A. 98 (10): 5538–43. doi:10.1073/pnas.091603098. PMC 33248
 . PMID 11331762.
. PMID 11331762. - 1 2 Zeidler M, Kleer CG (2006). "The Polycomb group protein Enhancer of Zeste 2: its links to DNA repair and breast cancer". J. Mol. Histol. 37 (5-7): 219–23. doi:10.1007/s10735-006-9042-9. PMID 16855786.
- 1 2 Völkel P, Dupret B, Le Bourhis X, Angrand PO (2015). "Diverse involvement of EZH2 in cancer epigenetics". Am J Transl Res. 7 (2): 175–93. PMC 4399085
 . PMID 25901190.
. PMID 25901190. - ↑ Chang CJ, Hung MC (2012). "The role of EZH2 in tumour progression". Br. J. Cancer. 106 (2): 243–7. doi:10.1038/bjc.2011.551. PMC 3261672
 . PMID 22187039.
. PMID 22187039. - ↑ Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM (2003). "EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells". Proc. Natl. Acad. Sci. U.S.A. 100 (20): 11606–11. doi:10.1073/pnas.1933744100. PMC 208805
 . PMID 14500907.
. PMID 14500907.
Further reading
- Kopecný J (1976). "[Rational approaches to the treatment of urinary incontinence (author's transl)]". Ceskoslovenská gynekologie. 41 (6): 408–9. PMID 975276.
- Price EA, Bourne SL, Radbourne R, Lawton PA, Lamerdin J, Thompson LH, Arrand JE (1998). "Rare microsatellite polymorphisms in the DNA repair genes XRCC1, XRCC3 and XRCC5 associated with cancer in patients of varying radiosensitivity". Somat. Cell Mol. Genet. 23 (4): 237–247. doi:10.1007/BF02674415. PMID 9542526.
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Narayana LS, Zhou ZQ, Adamson AW, Sorensen KJ, Chen DJ, Jones NJ, Thompson LH (1998). "XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages". Mol. Cell. 1 (6): 783–793. doi:10.1016/S1097-2765(00)80078-7. PMID 9660962.
- Pierce AJ, Johnson RD, Thompson LH, Jasin M (1999). "XRCC3 promotes homology-directed repair of DNA damage in mammalian cells". Genes Dev. 13 (20): 2633–2638. doi:10.1101/gad.13.20.2633. PMC 317094
 . PMID 10541549.
. PMID 10541549. - Winsey SL, Haldar NA, Marsh HP, Bunce M, Marshall SE, Harris AL, Wojnarowska F, Welsh KI (2000). "A variant within the DNA repair gene XRCC3 is associated with the development of melanoma skin cancer". Cancer Res. 60 (20): 5612–6. PMID 11059748.
- Kurumizaka H, Ikawa S, Nakada M, Eda K, Kagawa W, Takata M, Takeda S, Yokoyama S, Shibata T (2001). "Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C". Proc. Natl. Acad. Sci. U.S.A. 98 (10): 5538–5543. doi:10.1073/pnas.091603098. PMC 33248
 . PMID 11331762.
. PMID 11331762. - Masson JY, Stasiak AZ, Stasiak A, Benson FE, West SC (2001). "Complex formation by the human RAD51C and XRCC3 recombination repair proteins". Proc. Natl. Acad. Sci. U.S.A. 98 (15): 8440–8446. doi:10.1073/pnas.111005698. PMC 37455
 . PMID 11459987.
. PMID 11459987. - Miller KA, Yoshikawa DM, McConnell IR, Clark R, Schild D, Albala JS (2002). "RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51". J. Biol. Chem. 277 (10): 8406–8411. doi:10.1074/jbc.M108306200. PMID 11744692.
- Liu N, Schild D, Thelen MP, Thompson LH (2002). "Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells". Nucleic Acids Res. 30 (4): 1009–1015. doi:10.1093/nar/30.4.1009. PMC 100342
 . PMID 11842113.
. PMID 11842113. - Shen H, Sturgis EM, Dahlstrom KR, Zheng Y, Spitz MR, Wei Q (2002). "A variant of the DNA repair gene XRCC3 and risk of squamous cell carcinoma of the head and neck: a case-control analysis". Int. J. Cancer. 99 (6): 869–872. doi:10.1002/ijc.10413. PMID 12115490.
- Duan Z, Shen H, Lee JE, Gershenwald JE, Ross MI, Mansfield PF, Duvic M, Strom SS, Spitz MR, Wei Q (2002). "DNA repair gene XRCC3 241Met variant is not associated with risk of cutaneous malignant melanoma". Cancer Epidemiol. Biomarkers Prev. 11 (10 Pt 1): 1142–3. PMID 12376526.
- Seedhouse C, Bainton R, Lewis M, Harding A, Russell N, Das-Gupta E (2003). "The genotype distribution of the XRCC1 gene indicates a role for base excision repair in the development of therapy-related acute myeloblastic leukemia". Blood. 100 (10): 3761–3766. doi:10.1182/blood-2002-04-1152. PMID 12393447.
- Smith TR, Miller MS, Lohman K, Lange EM, Case LD, Mohrenweiser HW, Hu JJ (2003). "Polymorphisms of XRCC1 and XRCC3 genes and susceptibility to breast cancer". Cancer Lett. 190 (2): 183–190. doi:10.1016/S0304-3835(02)00595-5. PMID 12565173.
- Jacobsen NR, Nexø BA, Olsen A, Overvad K, Wallin H, Tjønneland A, Vogel U (2004). "No association between the DNA repair gene XRCC3 T241M polymorphism and risk of skin cancer and breast cancer". Cancer Epidemiol. Biomarkers Prev. 12 (6): 584–5. PMID 12815008.
- Zhu G, Duffy DL, Turner DR, Ewen KR, Montgomery GW, Martin NG (2005). "Linkage and association analysis of radiation damage repair genes XRCC3 and XRCC5 with nevus density in adolescent twins". Twin Research. 6 (4): 315–321. doi:10.1375/136905203322296683. PMID 14511439.
- Bertram CG, Gaut RM, Barrett JH, Randerson-Moor J, Whitaker L, Turner F, Bataille V, dos Santos Silva I, Swerdlow AJ, Bishop DT, Newton Bishop JA (2004). "An assessment of a variant of the DNA repair gene XRCC3 as a possible nevus or melanoma susceptibility genotype". J. Invest. Dermatol. 122 (2): 429–432. doi:10.1046/j.0022-202X.2003.12541.x. PMID 15009726.
- Tarsounas M, Davies AA, West SC (2004). "RAD51 localization and activation following DNA damage". Philos. Trans. R. Soc. Lond., B, Biol. Sci. 359 (1441): 87–93. doi:10.1098/rstb.2003.1368. PMC 1693300
 . PMID 15065660.
. PMID 15065660.