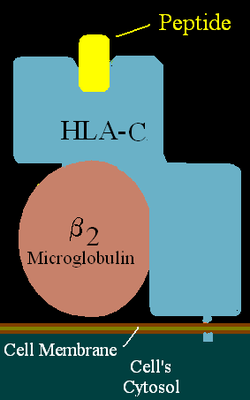
HLA-C
| View/Edit Human | |||
HLA-C belongs to the MHC (human = HLA) class I heavy chain receptors. The C receptor is a heterodimer consisting of a HLA-C mature gene product and β2-microglobulin. The mature C chain is anchored in the membrane. MHC Class I molecules, like HLA-C, are expressed in nearly all cells, and present small peptides to the immune system which surveys for non-self peptides.
HLA-C is a locus on chromosome 6, which encodes for a large number of HLA-C alleles that are Class-I MHC receptors. HLA-C, localized proximal to the HLA-B locus, is located on the distal end of the HLA region. Most HLA C:B haplotypes are in strong linkage disequilibrium and many are as ancient as the human species itself.
Disease associations
By serotype
Cw1: Multinodular goiters[3]
By allele
C*16: B chronic lymphocytic leukemia[4]
Nomenclature
C*01
- Cw1 Serotype: C*01:02 and C*01:09
- Cw11 ?
- C*01:04 to *01:08
C*02
- Cw2 Serotype: C*02:02 and *02:08
- C*02:03 to *02:07, and 02:09
C*03
- Cw9 Serotype: C*03:03
- Cw10 Serotype: C*03:02, *03:04, and *03:06
- Cw3 Serotype: C*03:07
- C*03:05 and 03:08
C*04
- Cw4 Serotype: C*0401, *0407, and *0410
C*05
- Cw5 Serotype: C*05:01 and *05:02
- C*05:03 to *05:06 and *05:08 to *05:10
C*06
- Cw6 Serotype: C*06:02 and *06:05
- C*06:03, *06:04 and *06:06 to *06:11
- Cw7 Serotype: C*07:01 to *07:06, *07:12, *07:14, *07:16
- C*07:07 to *07:11, *07:13, *07:15, and *07:17 to *07:29
C*08
- Cw8 Serotype: C*08:01, *08:02 and *08:03
- C*08:05 to *08:12
C*12:02 to *12:15
C*14:02 to *14:05
C*15:01 to *15:11
C*16:01 to *16:06
C*17:01 to *17:03
C*18:01 and *18:02
Common Haplotype
Cw4-B35 (Western Africa to Native Americans) Cw7-B7 (Western Eurasia, South Africa) Cw7-B8 (Western Eurasia) Cw1-B46 (China, Indochina) Cw5-B44 (Western Eurasia)
Interactions
HLA-C has been shown to interact with:
- KIR2DL1,[5][6][7][8] with various members of the
- LILRA3 [9] and
- Leukocyte immunoglobulin-like receptor family, particularly the activating receptors LILRA1 and
References
- ↑ "Diseases that are genetically associated with HLA-C view/edit references on wikidata".
- ↑ "Human PubMed Reference:".
- ↑ Ríos A, Rodríguez JM, Moya MR, Galindo PJ, Canteras M, Alvarez MR, Parrilla P (2006). "Associations of HLA-C alleles with multinodular goiters: study in a population from southeastern Spain.". Arch Surg. 141 (2): 123–8. doi:10.1001/archsurg.141.2.123. PMID 16490887.
- ↑ Montes-Ares O, Moya-Quiles MR, Montes-Casado M, Guerra-Pérez N, Campillo JA, González C, López-Bermejo A, Tamayo M, Majado MJ, Parrado A, Muro M, Marín L, Alvarez-López MR (2006). "Human leucocyte antigen-C in B chronic lymphocytic leukaemia.". Br J Haematol. 135 (4): 517–9. doi:10.1111/j.1365-2141.2006.06334.x. PMID 17054674.
- ↑ Boyson JE, Erskine R, Whitman MC, Chiu M, Lau JM, Koopman LA, Valter MM, Angelisova P, Horejsi V, Strominger JL (December 2002). "Disulfide bond-mediated dimerization of HLA-G on the cell surface". Proc. Natl. Acad. Sci. U.S.A. 99 (25): 16180–5. doi:10.1073/pnas.212643199. PMC 138585
 . PMID 12454284.
. PMID 12454284. - ↑ Baba E, Erskine R, Boyson JE, Cohen GB, Davis DM, Malik P, Mandelboim O, Reyburn HT, Strominger JL (December 2000). "N-linked carbohydrate on human leukocyte antigen-C and recognition by natural killer cell inhibitory receptors". Hum. Immunol. 61 (12): 1202–18. doi:10.1016/s0198-8859(00)00184-1. PMID 11163076.
- ↑ Valés-Gómez M, Reyburn HT, Mandelboim M, Strominger JL (September 1998). "Kinetics of interaction of HLA-C ligands with natural killer cell inhibitory receptors". Immunity. 9 (3): 337–44. doi:10.1016/s1074-7613(00)80616-0. PMID 9768753.
- ↑ Fan QR, Long EO, Wiley DC (May 2001). "Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex". Nat. Immunol. 2 (5): 452–60. doi:10.1038/87766. PMID 11323700.
- ↑ Jones DC, Kosmoliaptsis V, Apps R, Lapaque N, Smith I, Kono A, Chang C, Boyle LH, Taylor CJ, Trowsdale J, Allen RL (March 2011). "HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding". J. Immunol. 186 (5): 2990–7. doi:10.4049/jimmunol.1003078. PMID 21270408.
Further reading
- Geyer M, Fackler OT, Peterlin BM (2001). "Structure--function relationships in HIV-1 Nef.". EMBO Rep. 2 (7): 580–5. doi:10.1093/embo-reports/kve141. PMC 1083955
 . PMID 11463741.
. PMID 11463741. - Greenway AL, Holloway G, McPhee DA, Ellis P, Cornall A, Lidman M (2004). "HIV-1 Nef control of cell signalling molecules: multiple strategies to promote virus replication.". J. Biosci. 28 (3): 323–35. doi:10.1007/BF02970151. PMID 12734410.
- Bénichou S, Benmerah A (2003). "[The HIV nef and the Kaposi-sarcoma-associated virus K3/K5 proteins: "parasites"of the endocytosis pathway]". Med Sci (Paris). 19 (1): 100–6. doi:10.1051/medsci/2003191100. PMID 12836198.
- Leavitt SA, SchOn A, Klein JC, Manjappara U, Chaiken IM, Freire E (2004). "Interactions of HIV-1 proteins gp120 and Nef with cellular partners define a novel allosteric paradigm.". Curr. Protein Pept. Sci. 5 (1): 1–8. doi:10.2174/1389203043486955. PMID 14965316.
- Tolstrup M, Ostergaard L, Laursen AL, Pedersen SF, Duch M (2004). "HIV/SIV escape from immune surveillance: focus on Nef.". Curr. HIV Res. 2 (2): 141–51. doi:10.2174/1570162043484924. PMID 15078178.
- Joseph AM, Kumar M, Mitra D (2005). "Nef: "necessary and enforcing factor" in HIV infection.". Curr. HIV Res. 3 (1): 87–94. doi:10.2174/1570162052773013. PMID 15638726.
- Anderson JL, Hope TJ (2005). "HIV accessory proteins and surviving the host cell.". Current HIV/AIDS reports. 1 (1): 47–53. doi:10.1007/s11904-004-0007-x. PMID 16091223.